- Homepage
- >
- Search Page
- > Slider
- >
Mitigating the Impacts of Climate Change: How EU HPC Centres of Excellence Are Meeting the Challenge
“The past seven years are on track to be the seven warmest on record,” according to the World Meteorological Organization. Furthermore, the earth is already experiencing the extreme weather consequences of a warmer planet in the forms of record snow in Madrid, record flooding in Germany and record wildfires in Greece in 2021 alone. Although EU HPC Centres of Excellence (CoEs) help to address current societal challenges like the Covid-19 pandemic, you might wonder, what can the EU HPC CoEs do about climate change? For some CoEs, the answer is fairly obvious. However just as with Covid-19, the contributions of other CoEs may surprise you!
Given that rates of extreme weather events are already increasing, what can EU HPC CoEs do to help today? The Centre of Excellence in Simulation of Weather and Climate in Europe (ESiWACE) is optimizing weather and climate simulations for the latest HPC systems to be fast and accurate enough to predict specific extreme weather events. These increasingly detailed climate models have the capacity to help policy makers make more informed decisions by “forecasting” each decision’s simulated long-term consequences, ultimately saving lives. Beyond this software development, ESiWACE also supports the proliferation of these more powerful simulations through training events, large scale use case collaborations, and direct software support opportunities for related projects.
Even excepting extreme weather and long-term consequences, though, climate change has other negative impacts on daily life. For example the World Health Organization states that air pollution increases rates of “stroke, heart disease, lung cancer, and both chronic and acute respiratory diseases, including asthma.” The HPC and Big Data Technologies for Global Systems Centre of Excellence (HiDALGO) exists to provide the computational and data analytic environment needed to tackle global problems on this scale. Their Urban Air Pollution Pilot, for example, has the capacity to forecast air pollution levels down to two meters based on traffic patterns and 3D geographical information about a city. Armed with this information and the ability to virtually test mitigations, policy makers are then empowered to make more informed and effective decisions, just as in the case of HiDALGO’s Covid-19 modelling.
What does MAterials design at the eXascale have to do with climate change? Among other things, MaX is dramatically speeding up the search for materials that make more efficient, safer, and smaller lithium ion batteries: a field of study that has had little success despite decades of searching. The otherwise human intensive process of finding new candidate materials moves exponentially faster when conducted computationally on HPC systems. Using HPC also ensures that the human researchers can focus their experiments on only the most promising material candidates.
Continuing with the theme of materials discovery, did you know that it is possible to “capture” CO2 from the atmosphere? We already have the technology to take this greenhouse gas out of our air and put it back into materials that keep it from further warming the planet. These materials could even be a new source of fuel almost like a man-made, renewable oil. The reason this isn’t yet part of the solution to climate change is that it is too slow. In answer, the Novel Materials Discovery Centre of Excellence (NoMaD CoE) is working on finding catalysts to speed up the process of carbon capture. Their recent success story about a publication in Nature discusses how they have used HPC and AI to identify the “genes” of materials that could make efficient carbon-capture catalysts. In our race against the limited amount of time we have to prevent the worst impacts of climate change, the kind of HPC facilitated efficiency boost experienced by MaX and NoMaD could be critical.
Once one considers the need of efficiency, it starts to become clear what the Centre of Excellence for engineering applications EXCELLERAT might be able to offer. Like all of the EU HPC CoEs, EXCELLERAT is working to prepare software to run on the next generation of supercomputers. This preparation is vital because the computers will use a mixture of processor types and be organized in a variety of architectures. Although this variety makes the machines themselves more flexible and powerful, it also demands increased flexibility from the software that runs on them. For example, the software will need the ability to dynamically change how work is distributed among processors depending on what kind and how many a specific supercomputer has. Without this ability, the software will run at the same speed no matter how big, fast, or powerful the computer is: as if it only knows how to work with a team of 5 despite having a team of 20. Hence, EXCELLERAT is preparing engineering simulation software to adapt to working efficiently on any given machine. This kind of simulation software is making it possible to more rapidly design new airplanes for characteristics like a shape that has less drag/better fuel efficiency, less sound pollution, and easier recycling of materials when the plane is too old to use.
Another CoE using HPC efficiency to make our world more sustainable is the Centre of Excellence for Combustion (CoEC). Focused exclusively on combustion simulation, they are working to discover new non-carbon or low-carbon fuels and more sustainable ways of burning them. Until now, the primary barrier to this kind of research has been the computing limitations of HPC systems, which could not support realistically detailed simulations. Only with the capacity of the latest and future machines will researchers finally be able to run simulations accurate enough for practical advances.
Outside of the pursuit for more sustainable combustion, the Energy Oriented Centre of Excellence (EoCoE) is boosting the efficiency of entirely different energy sources. In the realm of Wind for Energy, their simulations designed for the latest HPC systems have boosted the size of simulated wind farms from 5 to 40 square kilometres, which allows researchers and industry to far better understand the impact of land terrain and wind turbine placement. They are also working outside of established wind energy technology to help design an entirely new kind of wind turbine.
In work also related to solar energy, the EoCoE Materials for Energy group is finding new materials to improve the efficiency of solar cells as well as separately working on materials to harvest energy from the mixture of salt and fresh water in estuaries. Meanwhile, the Water for Energy group is improving the modelling of ground water movement to enable more efficient positioning of geothermal wells and the Fusion for Energy group is working to improve the accuracy of models to predict fusion energy output.
EoCoE is also developing simulations to support Meteorology for Energy including the ability to predict wind and solar power capacity in Europe. Unlike our normal daily forecast, energy forecasts need to calculate the impact of fog or cloud thickness on solar cells and wind fluctuations caused by extreme temperature shifts or storms on wind turbines. Without this more advanced form of weather forecasting, it is unfeasible for these renewable but variable energy sources to make up a large amount of the power supplied to our fluctuation sensitive grids. Before we are able to rely on wind and solar power, it will be essential to predict renewable energy output in time to make changes or supplement with alternate energy sources, especially in light of the previously mentioned increase in extreme weather events.
Suffice it to say that climate change poses a variety of enormous challenges. The above describes only some of the work EU HPC CoEs are already doing and none of what they may be able to do in the future! For instance, HiDALGO also has a migration modelling program currently designed to help policy makers divert resources most effectively to migrations caused by conflict. However, similar principles could theoretically be employed in combination with weather modelling like that done by ESiWACE to create a climate migration model. Where expertise meets collaboration, the possibilities are endless! Make sure to follow the links above and our social media handles below to stay up to date on EU HPC CoE activities.
FocusCoE at EuroHPC Summit Week 2022
With the support of the FocusCoE project, almost all European HPC Centres of Excellence (CoEs) participated once again in the EuroHPC Summit Week (EHPCSW) this year in Paris, France: the first EHPCSW in person since 2019’s event in Poland. Hosted by the French HPC agency Grand équipement national de calcul intensif (GENCI), the conference was organised by Partnership for Advanced Computing in Europe (PRACE), the European Technology Platform for High-Performance Computing (ETP4HPC), The EuroHPC Joint Undertaking (EuroHPC JU), and the European Commission (EC).As usual, this year’s event gathered the main European HPC stakeholders from technology suppliers and HPC infrastructures to scientific and industrial HPC users in Europe.

At the workshop on the European HPC ecosystem on Tuesday 22 March at 14:45, where the diversity of the ecosystem was presented around the Infrastructure, Applications, and Technology pillars, project coordinator Dr. Guy Lonsdale from Scapos talked about FocusCoE and the CoEs’ common goal.
Later that day from 16:30 until 18:00h, the FocusCoE project hosted a session titled “European HPC CoEs: perspectives for a healthy HPC application eco-system and Exascale” involving most of the EU CoEs. The session discussed the key role of CoEs in the EuroHPC application pillar, focussing on their impact for building a vibrant, healthy HPC application eco-system and on perspectives for Exascale applications. As described by Dr. Andreas Wierse on behalf of EXCELLERAT, “The development is continuous. To prepare companies to make good use of this technology, it’s important to start early. Our task is to ensure continuity from using small systems up to the Exascale, regardless of whether the user comes from a big company or from an SME”.
Keen interest in the agenda was also demonstrated by attendees from HPC related academia and industry filling the hall to standing room only. In light of the call for new EU HPC Centres of Excellence and the increasing return to in-person events like EHPCSW, the high interest in preparing the EU for Exascale has a bright future.
AI Café: How can HPC technologies help AI
On March 17th, FocusCoE participated in a live AI for Media Web Café alongside the three Centres of Excellence: RAISE, CoEC, and HiDALGO. The virtual session brought together the CoEs working in AI sectors to explain how HPC technologies can help AI. In all, over 40 participants from industry and research joined the hour and a half café.
Starting off the presentations, Xavier Salazar introduced FocusCoE and the resources available at the “one stop shop” of our website such as technological offerings. Here, anyone from industry or research who wants to learn more can also read up on use cases, search available codes and software packages, and link directly to the CoEs of interest.
Next, the CoEs presented several case studies on how they are using AI in combination with HPC technologies to solve real-life problems. Although each CoE’s application of AI differed, some common themes emerged in answer to the question, “How can HPC help AI?” Firstly, AI is now benefitting from the increasing availability of large and even “big” data sets but often can’t use them in their entirety due to excessive processing time. This is by far the clearest example of how HPC can help. In a use case described by Andreas Lintermann on behalf of CoE RAISE, a dataset that was estimated to take over 300 hours to process using 4 GPUs was modified to run on HPC systems theoretically as large as 2000 GPUs in as little as 45 minutes! With the ability to more quickly train AI models using more data, it is also possible to increase the accuracy of the resulting models or surrogates. In turn, building more accurate surrogates speeds up the ability to run accurate simulations since one no longer needs to build the simulation models by hand.
Using AI to build data model surrogates also has benefits for data privacy, as discussed by Christoph Schweimer from HiDALGO. When modelling how messages spread across social media, researchers initially had to build social network graphs manually from data harvested from real social media users, whose privacy had to be strictly protected. However, with HPC computing resources, HiDALGO researchers were able to use those real graphs to train AI to build simulated social network graphs instead. These simulated graphs share the same characteristics of real graphs but require far less time to create and don’t rely on any real-user data: thus holding no privacy risks to users.
The experience gained through these use cases has naturally brought several opportunities and challenges to light, which were also discussed over the course of the program. For instance, Temistocle Grenga from CoEC highlighted the existing bottleneck of moving data between different types of processors (CPU and GPU, as examples).
Lastly, CoEs summarized the numerous resources in terms of services and training opportunities they provide to help AI experts learn to exploit the benefits of HPC. As an immediate example, CoEC will participate this week in South-East Europe Combustion Spring School 2022. For ongoing information on training like this, make sure to bookmark our training calendar, which shows events from all the EU HPC CoEs.
For the full recording of this event, check out the video below!
FocusCoE Hosts Intel OneAPI Workshop for the EU HPC CoEs
On March 2, 2022 FocusCoE hosted Intel for a workshop introducing the oneAPI development environment. In all, over 40 researchers representing the EU HPC Centres of Excellence (CoEs)were able to attend the single day workshop to gain an overview of OneAPI. The 8 presenters from Intel gave presentations through the day covering the OneAPI vision, design, toolkits, a use case with GROMACS (which is already used by some of the EU HPC CoEs), and specific tools for migration and debugging.
Launched in 2019, the Intel OneAPI cross-industry, open, standards-based unified programming model is being designed to deliver a common developer experience across accelerator architectures. With the time saved designing for specific accelerators, OneAPI is intended to enable faster application performance, more productivity, and greater innovation. As summarized on Intel’s OneAPI website, “Apply your skills to the next innovation, and not to rewriting software for the next hardware platform.” Given the work that EU HPC CoEs are currently doing to optimise codes for Exascale HPC systems, any tools that make this process faster and more efficient can only boost CoEs capacity for innovation and preparedness for future heterogeneous systems.
The OneAPI industry initiative is also encouraging collaboration on the oneAPI specification and compatible oneAPI implementations. To that end, Intel is investing time and expertise into events like this workshop to give researchers the knowledge they need not only to use but help improve OneAPI. The presenters then also make themselves available after the workshop to answer questions from attendees on an ongoing basis. Throughout our event, participants were continuously able to ask questions and get real-time answers as well as offers for further support from software architects, technical consulting engineers, and the researcher who presented a use case. Lastly, the full video and slides from presentations are available below for any CoEs who were unable to attend or would like a second look at the detailed presentations.

CoEs at Teratec Forum 2021 and ISC21
With the support of FocusCoE, a number of HPC CoEs will give short presentations at the virtual PRACE booth in the following two HPC-related events: Teratec Forum 2021 and ISC2021 that will take place towards the end of this month. See the schedule below for more details. Please reserve the slots in your calendars, registration details will be provided on the PRACE website soon!
“We are happy to see that FocusCoE was able to help the HPC CoEs to have a significant presence at this year’s editions of ISC and Teratec Forum, two major HPC events, enabled through our good synergies with PRACE”, says Guy Lonsdale, FocusCoE coordinator.
Teratec Forum 2021 schedule
Date / Event | Time slot CEST | Title | Speaker | Organisation |
Tue 22 June | 11:00 – 11:15 | EoCoE-II: Towards exascale for Energy | Edouard Audit, EoCoE-II coordinator | CEA (France) |
| 14:30 – 14:45 | POP CoE: Free Performance Assessments for the HPC Community | Bernd Mohr | Jülich Supercomputing Centre |
Thu 24 June | 13:45 – 14:00 | EXCELLERAT – paving the way for the evolution towards Exascale | Amgad Dessoky / Sophia Honisch | HLRS |
ISC 2021 schedule
Date / Event | Time slot CEST | Title | Speaker | Organisation |
Thu 24 June | 13:45 – 14:00 | EXCELLERAT – paving the way for the evolution towards Exascale | Amgad Dessoky / Sophia Honisch | HLRS |
Fri 25 June | 11:00 – 11:15 | The Center of Excellence for Exascale in Solid Earth (ChEESE) | Alice-Agnes Gabriel | Geophysik, University of Munich |
| 15:30 – 15:45 | EoCoE-II: Towards exascale for Energy | Edouard Audit, EoCoE-II coordinator | CEA (France) |
Tue 29 June | 11:00 – 11:15 | Towards a maximum utilization of synergies of HPC Competences in Europe | Bastian Koller, HLRS | HLRS |
Wed 30 June | 10:45 -11:00 | CoE | Dr.-Ing. Andreas Lintermann | Jülich Supercomputing Centre, Forschungszentrum Jülich GmbH |
Thu 1 July | 11:00 -11:15 | POP CoE: Free Performance Assessments for the HPC Community | Bernd Mohr | Jülich Supercomputing Centre |
| 14:30 -14:45 | TREX: an innovative view of HPC usage applied to Quantum Monte Carlo simulations | Anthony Scemama (1), William Jalby (2), Cedric Valensi (2), Pablo de Oliveira Castro (2) | (1) Laboratoire de Chimie et Physique Quantiques, CNRS-Université Paul Sabatier, Toulouse, France (2) Université de Versailles St-Quentin-en-Yvelines, Université Paris Saclay, France |
Please register to the short presentations through the PRACE event pages here:
| PRACE Virtual booth at Teratec Forum 2021 | PRACE Virtual booth at ISC2021 |
| prace-ri.eu/event/teratec-forum-2021/ | prace-ri.eu/event/praceisc-2021/ |
HiDALGO success story: Assisting decision makers to solve Global Challenges with HPC applications – Covid-19 modelling
Organisations & Codes Involved:
The National Health Service (NHS) oversees offering public health services in United Kingdom. It is now dealing with the COVID-19 pandemic.
Brunel University London is a dynamic institution that plays a significant role in the higher education sector. It carries out applied research on different topics, such as software engineering, intelligent data analysis, human computer interaction, information systems, and systems biology.
CHALLENGE:
The current pandemic situation has increased the NHS need of supporting tools to detect, predict and even prevent the virus spread behaviour. Knowing in advance this information will support them to take the appropriate decisions while considering health and care capabilities. In addition, the advance warning of new pandemic waves (or when they may subside) can help health authorities to rescale the capacity for non-urgent care, and ensure the timely arrangement of surge intensive-care capacity.
SOLUTION:
To tackle these challenges HiDALGO developed a tool: FACS, the Flu and Coronavirus Simulator, which is an agent-based model that also incorporates SEIRDI (Susceptible-Exposed-Infectious-Recovered-Dead-Immunized) states for all agents.
FACS approximates viral spread on the individual building level, and incorporates geospatial data sources from OpenStreetMap. In this way COVID-19 spread is modelled at local level, providing estimations of the spread of infections and hospital arrivals, given a range of public health interventions. Lastly, FACS supports the modelling of vaccination policies, as well as the introduction of new viral strains and changes in vaccine efficacy.
SOCIETAL & ECONOMIC IMPACT:
Although it is not a market topic itself, global challenges have been gaining importance in the last years, especially those related to climate change but also others such as peace and conflict. HiDALGO offers a set of tools, services, datasets and resources to define models that may predict situations under certain scenarios that can influence any decision to be taken.
More specifically, this tool FACS provides the needed information to decision makers so they can set up the appropriate measure and provide the necessary means at any time.
Indeed the tool helps the NHS to identify peaks of contagion in order to avoid sanitary collapses. Taking the appropriate decisions at the right moment represents a better investment of public resources and, what is more important, saving lives. Moreover, it supports to make better decisions and at appropriate time, to limit the problematic economic consequences of lockdowns and of the other measures taken in pandemic times.
BENEFITS FOR FURTHER RESEARCH:
- Support for the preparatory efforts by the health service for the second and third waves of the pandemic in West London.
- Better understanding of the nature of the current situation and the effect of different measures, such as lockdowns and vaccine efficacy levels.
- Provide models and elements about the foreseen evolution to limit the problematic economic consequences of the lockdowns and the various limitations due to the pandemic.
HiDALGO success story: Assisting decision makers to solve Global Challenges with HPC applications – Migration issues
Organisations & Codes Involved:
Save the Children is an NGO promoting policy changes to gain more rights for young people, especially by enforcing the UN Declaration of the Rights of the Child.
Brunel University London is a dynamic institution that plays a significant role in the higher education sector. It carries out applied research on different topics, such as software engineering, intelligent data analysis, human computer interaction, information systems, and systems biology.
CHALLENGE:
At the same time of the current COVID-19 pandemic, other crises have not stopped like forced migration due to conflicts. In fact, the number of forcibly displaced people is still very high, with over 70 million persons being forced to leave their homes. Save the Children provides support in these countries and needs more accurate estimations on people flows and even destinations to send the appropriate amount of help to the right place.
SOLUTION:
To tackle these challenges HiDALGO provides the tool Flee 2.0, a forced migration model which includes refugees and internally displaced people. It places agents that represent displaced persons in areas of conflict and uses movement rules to mimic their behavior as they attempt to find safety.
The code extracts location and route data from OpenStreetMap, and can be validated against UNHCR data for historical conflicts. As output, Flee provides a forecast of the amount (and location) of people that can be displaced given different conflict development scenarios.
SOCIETAL & ECONOMIC IMPACT:
Although it is not a market topic itself, global challenges have been gaining importance in the last years, especially those related to climate change but also others such as peace and conflict. HiDALGO offers a set of tools, services, datasets and resources to define models that may predict situations under certain scenarios that can influence any decision to be taken.
More specifically, Flee 2.0 provides the needed information to decision makers so they can set up the appropriate measure and provide the necessary means at any time.
For instance Flee 2.0 is used to estimate the expected number of refugee arrivals when conflicts occur in North Ethiopia, and to investigate how different conflict developments could affect the number of arrivals. The model development is partially guided by on-site experts and typically provides forecasts of approximately 3 month duration. Although work is ongoing on this project and the models are still basic, the aim is to establish a systematically developed mathematical model to improve the understanding of Save the Children about the migration situation, and support the preparation to mitigate the humanitarian impact of a potential upcoming crisis.
BENEFITS FOR FURTHER RESEARCH:
- Accurate predictions of where people may arrive in Sudan and how quicky, if new violence occurs in North
- Estimate of the effect of different conflict developments on the expected number of arrivals.
- Estimate of the effect of specific policy decisions on the expected number of arrivals.
Watch The Presentations Of The First CoE Joint Technical Workshop

Watch the recordings of the presentations from the first technical CoE workshop. The virtual event was organized by the three HPC Centres of Excellence ChEESE, EXCELLRAT and HiDALGO. The agenda for the workshop was structured in these four session:
Session 1: Load balancing
Session 2: In situ and remote visualisation
Session 3: Co-Design
Session 4: GPU Porting
You can also download a PDF version for each of the recorded presentations. The workshop took place on January 27 – 29, 2021.
Session 1: Load balancing
Title: Introduction by chairperson
Speaker: Ricard Borell (BSC)
Title: Load balancing strategies used in AVBP
Speaker: Gabriel Staffelbach (CERFACS)
Title: Addressing load balancing challenges due to fluctuating performance and non-uniform workload in SeisSol and ExaHyPE
Speaker: Michael Bader (TUM)
Title: On Discrete Load Balancing with Diffusion Type Algorithms
Speaker: Robert Elsäßer (PLUS)
Session 2: In situ and remote visualisation
Title: Introduction by chairperson
Speaker: Lorenzo Zanon & Anna Mack (HLRS)
Title: An introduction to the use of in-situ analysis in HPC
Speaker: Miguel Zavala (KTH)
Title: In situ visualisation service in Prace6IP
Speaker: Simone Bnà (CINECA)
Title: Web-based Visualisation of air pollution simulation with COVISE
Speaker: Anna Mack (HLRS)
Title: Virtual Twins, Smart Cities and Smart Citizens
Speaker: Leyla Kern, Uwe Wössner, Fabian Dembski (HLRS)
Session 3: Co-Design
Title: Introduction by chairperson, and Excellerat’s Co-Design Methodology
Speaker: Gavin Pringle (EPCC)
Title: Accelerating codes on reconfigurable architectures
Speaker: Nick Brown (EPCC)
Title: Benchmarking of Current Architectures for Improvements
Speaker: Nikela Papadopoulou (ICCS)
Title: Example Co-design Approach with the Seissol and Specfem3D Practical cases
Speaker: Georges-Emmanuel Moulard (ATOS)
Title: Exploitation of Exascale Systems for Open-Source Computational Fluid Dynamics by Mainstream Industry
Speaker: Ivan Spisso (CINECA)
Session 4: GPU Porting
Title: Introduction
Speaker: Giorgio Amati (CINECA)
Title: GPU Porting and strategies by ChEESE
Speaker: Piero Lanucara (CINECA)
Title: GPU porting by third party library
Speaker: Simone Bnà (CINECA)
How EU projects work on supercomputing applications to help contain the corona virus pandemic
The Centres of Excellence in high-performance computing are working to improve supercomputing applications in many different areas: from life sciences and medicine to materials design, from weather and climate research to global system science. A hot topic that affects many of the above-mentioned areas is, of course, the fight against the corona virus pandemic.
There are rather obvious challenges for those EU projects that are developing HPC applications for simulations in medicine or in the life sciences, like CompBioMed (Biomedicine) BioExcel (Biomolecular Research), and PerMedCoE (Personalized Medicine). But also other projects from scientific areas, that you would, at first sight, not directly relate to research on the pandemic, are developing and using appropriate applications to model the virus and its spread, and support policy makers with computing-heavy simulations. For example, did you know that researchers can simulate the possible spread of the virus on a local level, taking into account measures like closing shops or quarantining residents?
This article gives an overview over the various ways in which EU projects are using supercomputing applications to tackle and support the global challenge of containing the pandemic.
Simulations for better and faster drug development
 CompBioMed is an EU-funded project working on applications for computational biomedicine. It is part of a vast international consortium across Europe and USA working on urgent coronavirus research. “Modelling and simulation is being used in all aspects of medical and strategic actions in our fight against coronavirus. In our case, it is being harnessed to narrow down drug targets from billions of candidate molecules to a handful that can be clinically trialled”, says Peter Coveney from University College London (UCL) who is heading CompBioMed’s efforts in this collaboration. The goal is to accelerate the development of antiviral drugs by modelling proteins that play critical roles in the virus life cycle in order to identify promising drug targets.
CompBioMed is an EU-funded project working on applications for computational biomedicine. It is part of a vast international consortium across Europe and USA working on urgent coronavirus research. “Modelling and simulation is being used in all aspects of medical and strategic actions in our fight against coronavirus. In our case, it is being harnessed to narrow down drug targets from billions of candidate molecules to a handful that can be clinically trialled”, says Peter Coveney from University College London (UCL) who is heading CompBioMed’s efforts in this collaboration. The goal is to accelerate the development of antiviral drugs by modelling proteins that play critical roles in the virus life cycle in order to identify promising drug targets.
Secondly, for drug candidates already being used and trialled, the CompBioMed scientists are modelling and analysing the toxic effects that these drugs may have on the heart, using supercomputing resources required to run simulations on such scales. The goal is to assess the drug dosage and potential interactions between drugs to provide guidance for their use in the clinic.
Finally, the project partners analysed a model used to inform the UK Government’s response to the pandemic. It has been found to contain a large degree of uncertainty in its predictions, leading it to seriously underestimate the first wave. “Epidemiological modelling has been and continues to be used for policy-making by governments to determine healthcare interventions”, says Coveney. “We have investigated the reliability of such models using HPC methods required to truly understand the uncertainty and sensitivity of these models.” As a conclusion, a better public understanding of the inherent uncertainty of models predicting COVID-19 mortality rates is necessary, saying they should be regarded as “probabilistic” rather than being relied upon to produce a particular and specific outcome.
Researchsquare: Model uncertainty and decision making: Predicting the Impact of COVID-19 Using the CovidSim Epidemiological Code
Arxiv.org: Scalable HPC and AI Infrastructure for COVID-19 Therapeutics
Arxiv.org: IMPECCABLE: Integrated Modeling PipelinE for COVID Cure by Assessing Better LEads
BioExcel is an EU-funded project developing some of the most popular applications for modelling and simulations of biomolecular systems. Along with code development, the project builds training programmes to address competence gaps in extreme-scale scientific computing for beginners, advanced users and system maintainers.
is an EU-funded project developing some of the most popular applications for modelling and simulations of biomolecular systems. Along with code development, the project builds training programmes to address competence gaps in extreme-scale scientific computing for beginners, advanced users and system maintainers.
When COVID-19 struck, BioExcel launched a series of actions to support the community on SARS-CoV-2 research, with an extensive focus on facilitating collaborations, user support, and providing access to HPC resources at partner centers. BioExcel partnered with Molecular Sciences Software Institute to establish the COVID-19 Molecular Structure and Therapeutics Hub to allow researchers to deposit their data and review other group’s submissions as well.
During this period, there was an urgent demand for diagnostics and sharing of data for COVID-19 applications had become vital more than ever. A dedicated BioExcel-CV19 web-server interface was launched to provide access to study molecules involved in the COVID-19 disease. This allowed the project to be a part of open access initiative promoted by the scientific community to make research accessible.
Recently, BioExcel endorsed the EU manifesto for COVID-19 Research launched by European Commission as part of their response to the coronavirus outbreak.
Modelling the electronic structure of the protease
 MaX (MAterials design at the eXascale) is a European Centre of Excellence aiming at materials modelling, simulations, discovery and design on the exascale supercomputing architectures.
MaX (MAterials design at the eXascale) is a European Centre of Excellence aiming at materials modelling, simulations, discovery and design on the exascale supercomputing architectures.
Though the main interest of the MaX flagship codes is then centered on materials science, the CoE is participating in the fight against SARS-CoV-2. Given the critical pandemic situation that the world is currently facing, an unprecedented effort is being devoted to the study of SARS-CoV-2 by researchers from different scientific communities and groups worldwide. From the biomolecular standpoint, particular focus is being devoted to the main protease, as well as to the spike protein. As such, it is an important potential antiviral drug target: if its function is inhibited, the virus remains immature and non-infectious. Using fragment-based screening, researchers have identified a number of small compounds that bind to the active site of the protease and can be used as a starting point for the development of protease inhibitors.
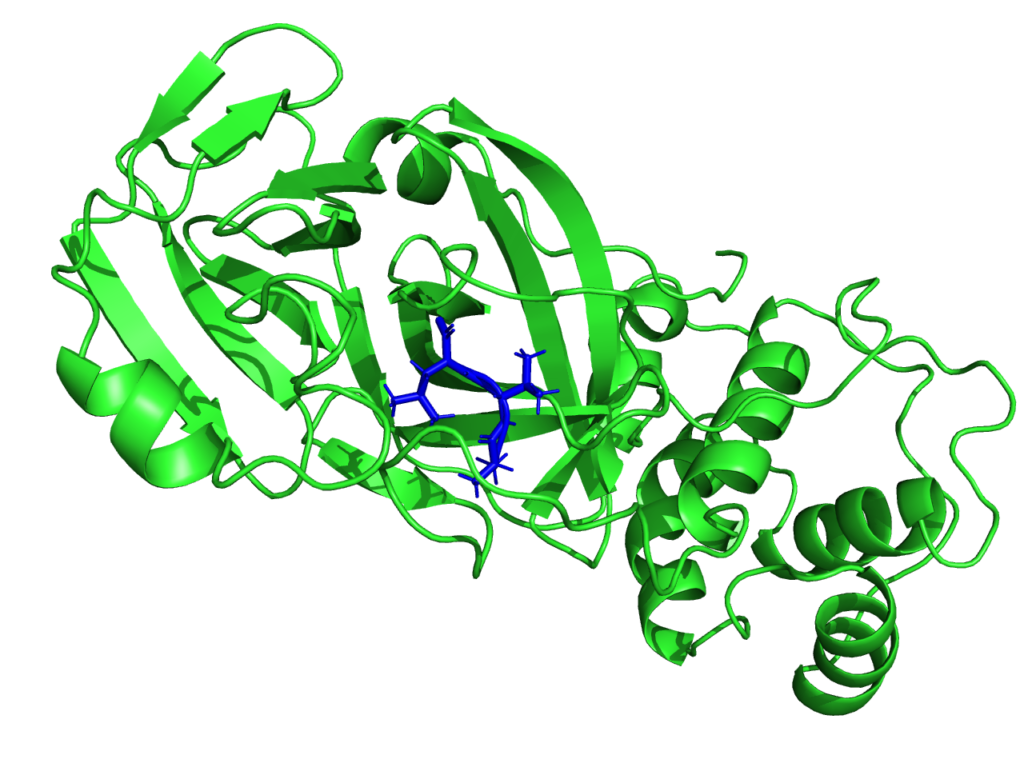
Among other quantities, MaX researchers now have the possibility to model the electronic structure of the protease in contact with a potential docked inhibitor, and provide new insights on the interactions between them by selecting specific amino-acids that are involved in the interaction and characterizing their polarities. This new approach proposed by the MaX scientists is complementary to the docking methods used up to now and based on in-silico research of the inhibitor. Biological systems are naturally composed of fragments such as amino-acids in proteins or nitrogenous bases in DNA.
With this approach, it is possible to evaluate whether the amino acid-based fragmentation is consistent with the electronic structure resulting from the QM computation. This is an important indicator for the end-user, as it enables to evaluate the quality of the information associated with a given fragment. Then, QM observables on the system’s fragments can be obtained, which are based on a population analysis of electronic density of the system, projected on the amino-acid.
A novelty that this approach enables is the possibility of quantifying the strength of the chemical interaction between the different fragments. It is possible to select a target region and identify which fragments of the systems interact with this region by sharing electrons with it.
“We can reconstruct the fragmentation of the system in such a way as to focus on an active site in a specific portion of the protein”, says Luigi Genovese from CEA (Commissariat à l’énergie atomique et aux énergies alternatives) who is heading Max’s efforts on this topic. “We think this modelling approach could inform efforts in protein design by granting access to variables otherwise impervious to observation.”
Improving drug design and biosensors
 The project E-CAM supports HPC simulations in industry and academia through software development, training and discussion in simulation and modeling. Project members are currently following two approaches to add to the research on the corona virus.
The project E-CAM supports HPC simulations in industry and academia through software development, training and discussion in simulation and modeling. Project members are currently following two approaches to add to the research on the corona virus.
Firstly, the SARS-CoV-2 virus that causes COVID-19 uses a main protease to be functional. One of the drug targets currently under investigation is an inhibitor for this protease. While efforts on simulations of binding stability and dynamics are being conducted, not much is known of the dynamical transitions of the binding-unbinding reaction. Yet, this knowledge is crucial for improved drug design. E-CAM aims to shed light on these transitions, using a software package developed by project teams at the University of Amsterdam and the Ecole Normale Superieure in Lyon.
Secondly, E-CAM contributes to the development of the software required to design a protein-based sensor for the quick detection of COVID-19. The sensor, developed at the partner University College Dublin with the initial purpose to target influenza, is now being adapted to SARS-CoV-2. This adaptation needs DNA sequences as an input for suitable antibody-epitope pairs. High-performance computing is required to identify these DNA sequences to design and simulate the sensors prior to their expression in cell lines, purification and validation.
Studying COVID-19 infections on the cell level
 The project PerMedCoE aims to optimise codes for cell-level simulations in high-performance computing, and to bridge the gap between organ and molecular simulations. The project started in October 2020.
The project PerMedCoE aims to optimise codes for cell-level simulations in high-performance computing, and to bridge the gap between organ and molecular simulations. The project started in October 2020.
“Multiscale modelling frameworks prove useful in integrating mechanisms that have very different time and space scales, as in the study of viral infection, human host cell demise and immune cells response. Our goal is to provide such a multiscale modelling framework that includes infection mechanisms, virus propagation and detailed signalling pathways,” says Alfonso Valencia, PerMedCoE project coordinator at the Barcelona Supercomputing Center.
The project team has developed a use case that focusses on studying COVID-19 infections using single-cell data. The work was presented to the research community at a specialized virtual conference in November, the Disease Map Community Meeting. “This use case is a priority in the first months of the project”, says Valencia.
On the technical level, disease maps networks will be converted to models of COVID-19 and human cells from the lung epithelium and the immune system. Then, the team will use omics data to personalise models of different patients’ groups, differentiated for example by age or gender. These data-tailored models will then be incorporated into a COVID-focussed version of the open source cell-level simulator PhysiCell.
Supporting policy makers and governments
 The HiDALGO project focusses on modelling and simulating the complex processes which arise in connection with major global challenges. The researchers have developed the Flu and Coronavirus Simulator (FACS) with the objective to support decision makers to provide an appropriate response to the current pandemic situation taking into account health and care capabilities.
The HiDALGO project focusses on modelling and simulating the complex processes which arise in connection with major global challenges. The researchers have developed the Flu and Coronavirus Simulator (FACS) with the objective to support decision makers to provide an appropriate response to the current pandemic situation taking into account health and care capabilities.
FACS is guided by the outcomes of SEIR (Susceptible-Exposed-Infectious-Recovered) models operating at national level. It uses geospatial data sources from Openstreet Map to approximate the viral spread in crowded places, while trading the potential routes to reach them.
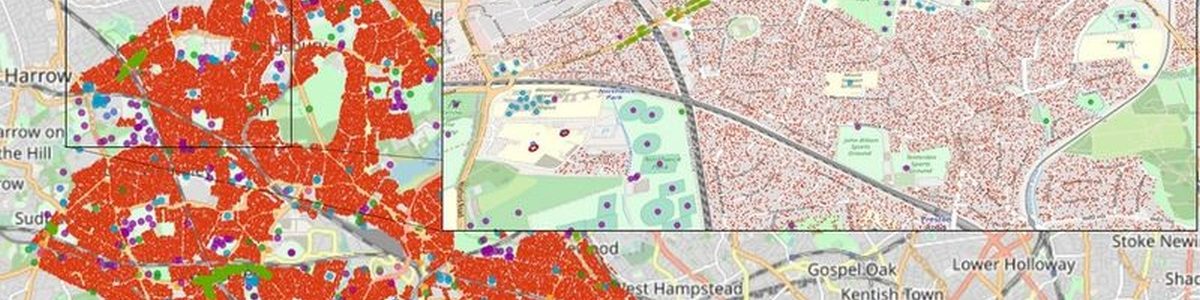
In this way, the simulator can model the COVID-19 spread at local level to provide estimations of infections and hospital arrivals, given a range of public health interventions, going from no interventions to lockdowns. Public authorities can use the results of the simulations to identify peaks of contagion, set appropriate measures to reduce spread and provide necessary means to hospitals to prevent collapses. “FACS has enabled us to forecast the spread of COVID-19 in regions such as the London Borough of Brent. These forecasts have helped local National Health Service Trusts to more effectively plan out health and care services in response to the pandemic.” says Derek Groen from the HiDALGO project partner Brunel University London.
Wired: Why London’s Tier 2 lockdown couldn’t be borough by borough
The Guardian: As other cities go into lockdown, why isn’t London having a second wave?
HiPEAC Magazine: Simulating COVID-19 andflu spread using HiDALGO
HiDALGO website: Pandemic Use Case
HiDALGO establishes a local flu and coronavirus detector
Cordis – EU research results: London calling: Simulation tool predicts second wave of COVID-19
 EXCELLERAT is a project that is usually focussing on supercomputing applications in the area of engineering. Nevertheless, a group of researchers from EXCELLERAT’s consortium partner SSC-Services GmbH, an IT service provider in Böblingen, Germany and the High-Performance Computing Center Stuttgart (HLRS) are also providing measures to contain the pandemic by supporting the German Federal Institute for Population Research (Bundesinstitut für Bevölkerungsforschung, BiB).
EXCELLERAT is a project that is usually focussing on supercomputing applications in the area of engineering. Nevertheless, a group of researchers from EXCELLERAT’s consortium partner SSC-Services GmbH, an IT service provider in Böblingen, Germany and the High-Performance Computing Center Stuttgart (HLRS) are also providing measures to contain the pandemic by supporting the German Federal Institute for Population Research (Bundesinstitut für Bevölkerungsforschung, BiB).
The scientists have developed an intelligent data transfer platform, which enables the BiB to upload data, perform computing-heavy simulations on the HLRS’ supercomputer Hawk, and download the results. The platform supports the work of BiB researchers in predicting the demand for intensive care units during the COVID-19 pandemic. “Nowadays, organisations face various issues while dealing with HPC calculations, HPC in general or even the access to HPC resources,” said Janik Schüssler, project manager at SSC Services. “In many cases, calculations are too complex and users do not have the required know-how with HPC technologies. This is the challenge that we have taken on. The BiB’s researchers had to access HLRS’s Hawk in a very complex way. With the help of our new platform, they can easily access Hawk from anywhere and run their simulations remotely.”
“This platform is part of EXCELLERAT’s overall strategy and tools development, which not only addresses the simulation part of engineering workflows, but provides users the necessary means to optimise their work”, said Bastian Koller, Project Coordinator of EXCELLERAT and HLRS’s Managing Director. “Extending the applicability of this platform to further use cases outside of the engineering domain is a huge benefit and increases the impact of the work performed in EXCELLERAT.”
ETP4HPC handbook 2020 released
The 2020 edition of the ETP4HPC Handbook of HPC projects is available. It offers a comprehensive overview over the European HPC landscape that currently consists of around 50 active projects and initiatives. Amongst these are the 14 Centres of Excellence and FocusCoE, that are also represented in this edition of the handbook.