- Homepage
- >
- Search Page
- > TREX
- >
FocusCoE at EuroHPC Summit Week 2022
With the support of the FocusCoE project, almost all European HPC Centres of Excellence (CoEs) participated once again in the EuroHPC Summit Week (EHPCSW) this year in Paris, France: the first EHPCSW in person since 2019’s event in Poland. Hosted by the French HPC agency Grand équipement national de calcul intensif (GENCI), the conference was organised by Partnership for Advanced Computing in Europe (PRACE), the European Technology Platform for High-Performance Computing (ETP4HPC), The EuroHPC Joint Undertaking (EuroHPC JU), and the European Commission (EC).As usual, this year’s event gathered the main European HPC stakeholders from technology suppliers and HPC infrastructures to scientific and industrial HPC users in Europe.

At the workshop on the European HPC ecosystem on Tuesday 22 March at 14:45, where the diversity of the ecosystem was presented around the Infrastructure, Applications, and Technology pillars, project coordinator Dr. Guy Lonsdale from Scapos talked about FocusCoE and the CoEs’ common goal.
Later that day from 16:30 until 18:00h, the FocusCoE project hosted a session titled “European HPC CoEs: perspectives for a healthy HPC application eco-system and Exascale” involving most of the EU CoEs. The session discussed the key role of CoEs in the EuroHPC application pillar, focussing on their impact for building a vibrant, healthy HPC application eco-system and on perspectives for Exascale applications. As described by Dr. Andreas Wierse on behalf of EXCELLERAT, “The development is continuous. To prepare companies to make good use of this technology, it’s important to start early. Our task is to ensure continuity from using small systems up to the Exascale, regardless of whether the user comes from a big company or from an SME”.
Keen interest in the agenda was also demonstrated by attendees from HPC related academia and industry filling the hall to standing room only. In light of the call for new EU HPC Centres of Excellence and the increasing return to in-person events like EHPCSW, the high interest in preparing the EU for Exascale has a bright future.
FocusCoE Hosts Intel OneAPI Workshop for the EU HPC CoEs
On March 2, 2022 FocusCoE hosted Intel for a workshop introducing the oneAPI development environment. In all, over 40 researchers representing the EU HPC Centres of Excellence (CoEs)were able to attend the single day workshop to gain an overview of OneAPI. The 8 presenters from Intel gave presentations through the day covering the OneAPI vision, design, toolkits, a use case with GROMACS (which is already used by some of the EU HPC CoEs), and specific tools for migration and debugging.
Launched in 2019, the Intel OneAPI cross-industry, open, standards-based unified programming model is being designed to deliver a common developer experience across accelerator architectures. With the time saved designing for specific accelerators, OneAPI is intended to enable faster application performance, more productivity, and greater innovation. As summarized on Intel’s OneAPI website, “Apply your skills to the next innovation, and not to rewriting software for the next hardware platform.” Given the work that EU HPC CoEs are currently doing to optimise codes for Exascale HPC systems, any tools that make this process faster and more efficient can only boost CoEs capacity for innovation and preparedness for future heterogeneous systems.
The OneAPI industry initiative is also encouraging collaboration on the oneAPI specification and compatible oneAPI implementations. To that end, Intel is investing time and expertise into events like this workshop to give researchers the knowledge they need not only to use but help improve OneAPI. The presenters then also make themselves available after the workshop to answer questions from attendees on an ongoing basis. Throughout our event, participants were continuously able to ask questions and get real-time answers as well as offers for further support from software architects, technical consulting engineers, and the researcher who presented a use case. Lastly, the full video and slides from presentations are available below for any CoEs who were unable to attend or would like a second look at the detailed presentations.

MEDICA Trade Fair 2021
On 17 November, 2021 Mariano Vasquez represented CompBioMed and PerMed CoE Centres of Excellence in an expert panel at the MEDICA Trade Fair’s Health IT Forum. Despite the challenges of the ongoing COVID-19 pandemic, the leading medical sector conference covering innovations in outpatient and clinical care was able to offer a 2021 hybrid conference with in-person attendance in Düsseldorf alongside virtual attendance from anywhere in the world. The Biomedical Experts Panel was comprised of professionals from imaging, data mining, data sharing, artificial intelligence, and robotics. Overall, they broadly discussed why IT solutions aren’t yet widely used in clinical settings and what the major barriers to wider adoption are.
In addition to the unanimously cited barrier of awareness, Mariano Vasquez explained how CompBioMed and PerMed Centres of Excellence are overcoming the specific difficulties in making clinicians aware of HPC’s potential in healthcare. “Work directly with doctors and let them take the reins of our research. When the clinicians are leading the way, they develop awareness of HPC and AI potential.” He continued by highlighting that, “in the CoEs we have resources to create a pipeline for using these tools,” Such as CompBioMed including computational biomedicine in curriculums for medical students.
Beyond awareness, all panel members indicated the further need for support from regulatory, legal, and clinical leadership stakeholders particularly in the case of the newest technologies like artificial intelligence. In the case of tools running off-site on HPC systems Mariano emphasized, “If you get the regulatory bodies saying the tools are ok, then the doctors and our customers like biomedical companies […]can start using the tools.”
Once an IT solution has awareness and hopefully broad stakeholder support, however, the next biggest challenge is training. In particular, Mariano highlighted how training for both clinicians and IT researchers is vital to building a common language for communication about IT solutions. “It’s like I have a hammer and the doctor has a nail and we need to link them. If the doctor’s never seen a hammer, he can’t know what to do with it even if the nail is in front of his nose.” When a researcher with perhaps a physics background shows something to a clinician, it is essential that the researcher understands the clinical problem, that both researcher and clinician can communicate about the solution in clinical terms, and that the clinician can provide feedback on the technology’s use.
In conclusion, all panel members made a call to unified action. As summarized by Mariano, “This is not science fiction, it is science […]This is something at hand now.” IT solutions exist but need support across stakeholder groups from researchers, to clinicians, through IT departments, and including leadership.
To the audience of health care professionals, Mariano encouraged, “Contact us. Don’t be shy!”
For more on the panel, log on and watch via the MEDICA On Demand Livestreams portal until 31 March, 2022.
For more on activities of the European Centres of Excellence such as CompBioMed and PerMed CoE, subscribe to our newsletter!
POP and PerMed Centers of Excellence are Getting Cell-Level Simulations Ready for Exascale
Success story # Highlights:
- Industry sector: Computational biology
- Key codes used: PhysiCell
- Keywords:
- Memory management
- Cell-cell interaction simulation
- OpenMP
- Good practice
Organisations & Codes Involved:



Technical Challenge:
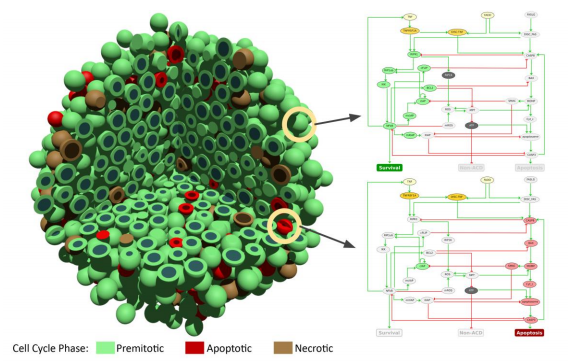
One of PerMedCoE’s use cases is the study of tumour evolution based on single-cell omics and imaging using PhysiCell. In order to simulate such large-scale problems that replicate real-world tumours, High Performance Computing (HPC) is essential. However, memory usage presents a challenge in HPC architectures and is one of the obstacles to optimizing simulations for running at Exascale.
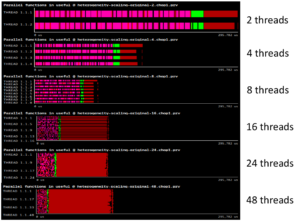
With a high number of threads computing in parallel, performance can be degraded because of concurrent memory allocations and deallocations. We observe that as the number of threads goes up, runtime is not reduced as expected. This is not a memory-bound problem, but a memory management one.
Solution:
Following the POP methodology and using the BSC tools (Extrae, Paraver, and BasicAnalysis), we determine that an overloading of a specific implementation of C++ operators is causing a high number of concurrent memory allocations and deallocations, causing the memory management library to perform synchronization and “steal” cycles of CPU from the running application.
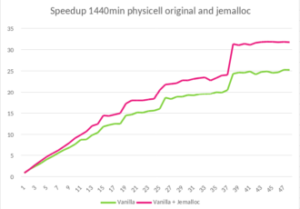
The proposal from POP is to avoid the overloading of operators that need allocation and deallocation of data structures. However, achieving this implies a major code change. To demonstrate the potential of the suggestion without doing the major code change, we suggest using an external library to improve the memory management: Jemalloc.
Jemalloc is “a general purpose malloc implementation that emphasizes fragmentation avoidance and scalable concurrency support” [1]. It can be integrated easily into any code by preloading the library, with LD_PRELOAD in Linux for example. After applying this solution to PhysiCell, we execute the same experiment and obtain a 1.45x speed up when using 48 OpenMP threads.
[1] cited from Jemalloc website. http://jemalloc.net
Business impact:
As the only transversal CoE, one of POP’s objectives is to advise and help the other HPC CoEs prepare their codes for the Exascale. This collaboration between POP and PerMed is a good example of the potential of these kinds of partnerships between CoEs.
POP provides the performance analysis expertise and tools, the experience of hundreds of codes analysed, and the best practices gathered from those analyses. PerMedCoE bring state of the art use cases and real-world problems to be solved. Together, they improve the performance and efficiency of cell-level agent-based simulation software: in this case PhysiCell.
One of the goals of PerMedCoE is the scaling-up of four different tools that address different types of simulations in personalized medicine and that were coded in different languages (C++, R, python, julia). These tools are being re-factored to scale up to Exascale. In this scaling-up, audits such as the ones POP can offer are essential to evaluate past developments and guide future ones.
Currently, only one of PerMed’s tools is able to use several nodes to run a single simulation, whereas the rest of the tools can only use all of the processors of a single node. Our scaling-up strategy is a heterogeneous one. We are implementing MPI on some tools, re-factoring others to other languages that can ease the use of HPC clusters such as Julia, or even targeting “many-task computing“ paradigms like in the case of model fitting.
The main motivation to have HPC versions of these simulation tools is to be able to simulate bigger, more complex cell-level agent-based models. Current models can obtain up to 10^6 cells, but it has been proven that most of the problems addressed in computational biology (cancerogenesis, cell lines growth, COVID-19 infection) usually target from 10^9 to 10^12 cells. In addition, most of these current simulations consider an over-simplified environment that is nowhere close to a real-life scenario. We aim to have complex multi-scale simulation frameworks that target these bigger, more complex simulations.
Benefits:
- Speedup of 1.45x on runtime with 48 threads in PhysiCell execution
- A good practice for High Performance C++ applications exported
- This behaviour in memory management systems will be detected more easily in future analysis
CoEs at Teratec Forum 2021 and ISC21
With the support of FocusCoE, a number of HPC CoEs will give short presentations at the virtual PRACE booth in the following two HPC-related events: Teratec Forum 2021 and ISC2021 that will take place towards the end of this month. See the schedule below for more details. Please reserve the slots in your calendars, registration details will be provided on the PRACE website soon!
“We are happy to see that FocusCoE was able to help the HPC CoEs to have a significant presence at this year’s editions of ISC and Teratec Forum, two major HPC events, enabled through our good synergies with PRACE”, says Guy Lonsdale, FocusCoE coordinator.
Teratec Forum 2021 schedule
Date / Event | Time slot CEST | Title | Speaker | Organisation |
Tue 22 June | 11:00 – 11:15 | EoCoE-II: Towards exascale for Energy | Edouard Audit, EoCoE-II coordinator | CEA (France) |
| 14:30 – 14:45 | POP CoE: Free Performance Assessments for the HPC Community | Bernd Mohr | Jülich Supercomputing Centre |
Thu 24 June | 13:45 – 14:00 | EXCELLERAT – paving the way for the evolution towards Exascale | Amgad Dessoky / Sophia Honisch | HLRS |
ISC 2021 schedule
Date / Event | Time slot CEST | Title | Speaker | Organisation |
Thu 24 June | 13:45 – 14:00 | EXCELLERAT – paving the way for the evolution towards Exascale | Amgad Dessoky / Sophia Honisch | HLRS |
Fri 25 June | 11:00 – 11:15 | The Center of Excellence for Exascale in Solid Earth (ChEESE) | Alice-Agnes Gabriel | Geophysik, University of Munich |
| 15:30 – 15:45 | EoCoE-II: Towards exascale for Energy | Edouard Audit, EoCoE-II coordinator | CEA (France) |
Tue 29 June | 11:00 – 11:15 | Towards a maximum utilization of synergies of HPC Competences in Europe | Bastian Koller, HLRS | HLRS |
Wed 30 June | 10:45 -11:00 | CoE | Dr.-Ing. Andreas Lintermann | Jülich Supercomputing Centre, Forschungszentrum Jülich GmbH |
Thu 1 July | 11:00 -11:15 | POP CoE: Free Performance Assessments for the HPC Community | Bernd Mohr | Jülich Supercomputing Centre |
| 14:30 -14:45 | TREX: an innovative view of HPC usage applied to Quantum Monte Carlo simulations | Anthony Scemama (1), William Jalby (2), Cedric Valensi (2), Pablo de Oliveira Castro (2) | (1) Laboratoire de Chimie et Physique Quantiques, CNRS-Université Paul Sabatier, Toulouse, France (2) Université de Versailles St-Quentin-en-Yvelines, Université Paris Saclay, France |
Please register to the short presentations through the PRACE event pages here:
| PRACE Virtual booth at Teratec Forum 2021 | PRACE Virtual booth at ISC2021 |
| prace-ri.eu/event/teratec-forum-2021/ | prace-ri.eu/event/praceisc-2021/ |
Video Of The Week: PerMedCoE: HPC Centre of Excellence on Personalised Medicine

HPC-enabled multiscale simulation helps uncover mechanistic insights of the SARS-CoV-2 infection
Short description
Results & Achievements
Objectives
Use Case Owner
Barcelona Supercomputing Center-Centro Nacional de Supercomputación (BSC-CNS)
How EU projects work on supercomputing applications to help contain the corona virus pandemic
The Centres of Excellence in high-performance computing are working to improve supercomputing applications in many different areas: from life sciences and medicine to materials design, from weather and climate research to global system science. A hot topic that affects many of the above-mentioned areas is, of course, the fight against the corona virus pandemic.
There are rather obvious challenges for those EU projects that are developing HPC applications for simulations in medicine or in the life sciences, like CompBioMed (Biomedicine) BioExcel (Biomolecular Research), and PerMedCoE (Personalized Medicine). But also other projects from scientific areas, that you would, at first sight, not directly relate to research on the pandemic, are developing and using appropriate applications to model the virus and its spread, and support policy makers with computing-heavy simulations. For example, did you know that researchers can simulate the possible spread of the virus on a local level, taking into account measures like closing shops or quarantining residents?
This article gives an overview over the various ways in which EU projects are using supercomputing applications to tackle and support the global challenge of containing the pandemic.
Simulations for better and faster drug development
 CompBioMed is an EU-funded project working on applications for computational biomedicine. It is part of a vast international consortium across Europe and USA working on urgent coronavirus research. “Modelling and simulation is being used in all aspects of medical and strategic actions in our fight against coronavirus. In our case, it is being harnessed to narrow down drug targets from billions of candidate molecules to a handful that can be clinically trialled”, says Peter Coveney from University College London (UCL) who is heading CompBioMed’s efforts in this collaboration. The goal is to accelerate the development of antiviral drugs by modelling proteins that play critical roles in the virus life cycle in order to identify promising drug targets.
CompBioMed is an EU-funded project working on applications for computational biomedicine. It is part of a vast international consortium across Europe and USA working on urgent coronavirus research. “Modelling and simulation is being used in all aspects of medical and strategic actions in our fight against coronavirus. In our case, it is being harnessed to narrow down drug targets from billions of candidate molecules to a handful that can be clinically trialled”, says Peter Coveney from University College London (UCL) who is heading CompBioMed’s efforts in this collaboration. The goal is to accelerate the development of antiviral drugs by modelling proteins that play critical roles in the virus life cycle in order to identify promising drug targets.
Secondly, for drug candidates already being used and trialled, the CompBioMed scientists are modelling and analysing the toxic effects that these drugs may have on the heart, using supercomputing resources required to run simulations on such scales. The goal is to assess the drug dosage and potential interactions between drugs to provide guidance for their use in the clinic.
Finally, the project partners analysed a model used to inform the UK Government’s response to the pandemic. It has been found to contain a large degree of uncertainty in its predictions, leading it to seriously underestimate the first wave. “Epidemiological modelling has been and continues to be used for policy-making by governments to determine healthcare interventions”, says Coveney. “We have investigated the reliability of such models using HPC methods required to truly understand the uncertainty and sensitivity of these models.” As a conclusion, a better public understanding of the inherent uncertainty of models predicting COVID-19 mortality rates is necessary, saying they should be regarded as “probabilistic” rather than being relied upon to produce a particular and specific outcome.
Researchsquare: Model uncertainty and decision making: Predicting the Impact of COVID-19 Using the CovidSim Epidemiological Code
Arxiv.org: Scalable HPC and AI Infrastructure for COVID-19 Therapeutics
Arxiv.org: IMPECCABLE: Integrated Modeling PipelinE for COVID Cure by Assessing Better LEads
BioExcel is an EU-funded project developing some of the most popular applications for modelling and simulations of biomolecular systems. Along with code development, the project builds training programmes to address competence gaps in extreme-scale scientific computing for beginners, advanced users and system maintainers.
is an EU-funded project developing some of the most popular applications for modelling and simulations of biomolecular systems. Along with code development, the project builds training programmes to address competence gaps in extreme-scale scientific computing for beginners, advanced users and system maintainers.
When COVID-19 struck, BioExcel launched a series of actions to support the community on SARS-CoV-2 research, with an extensive focus on facilitating collaborations, user support, and providing access to HPC resources at partner centers. BioExcel partnered with Molecular Sciences Software Institute to establish the COVID-19 Molecular Structure and Therapeutics Hub to allow researchers to deposit their data and review other group’s submissions as well.
During this period, there was an urgent demand for diagnostics and sharing of data for COVID-19 applications had become vital more than ever. A dedicated BioExcel-CV19 web-server interface was launched to provide access to study molecules involved in the COVID-19 disease. This allowed the project to be a part of open access initiative promoted by the scientific community to make research accessible.
Recently, BioExcel endorsed the EU manifesto for COVID-19 Research launched by European Commission as part of their response to the coronavirus outbreak.
Modelling the electronic structure of the protease
 MaX (MAterials design at the eXascale) is a European Centre of Excellence aiming at materials modelling, simulations, discovery and design on the exascale supercomputing architectures.
MaX (MAterials design at the eXascale) is a European Centre of Excellence aiming at materials modelling, simulations, discovery and design on the exascale supercomputing architectures.
Though the main interest of the MaX flagship codes is then centered on materials science, the CoE is participating in the fight against SARS-CoV-2. Given the critical pandemic situation that the world is currently facing, an unprecedented effort is being devoted to the study of SARS-CoV-2 by researchers from different scientific communities and groups worldwide. From the biomolecular standpoint, particular focus is being devoted to the main protease, as well as to the spike protein. As such, it is an important potential antiviral drug target: if its function is inhibited, the virus remains immature and non-infectious. Using fragment-based screening, researchers have identified a number of small compounds that bind to the active site of the protease and can be used as a starting point for the development of protease inhibitors.
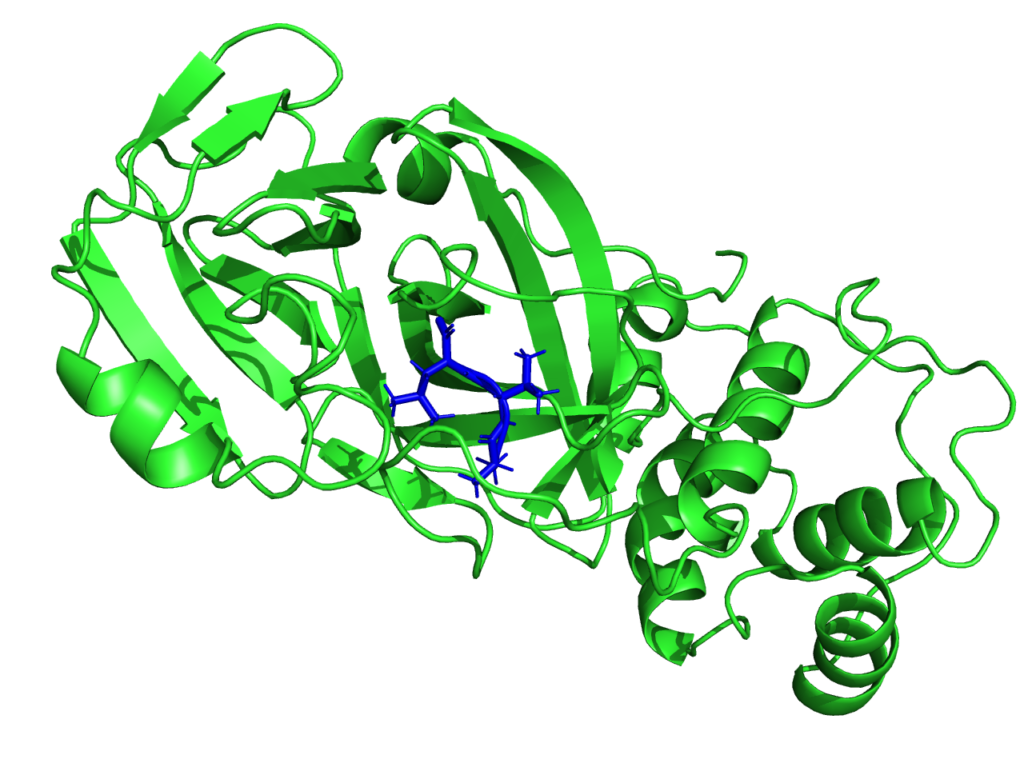
Among other quantities, MaX researchers now have the possibility to model the electronic structure of the protease in contact with a potential docked inhibitor, and provide new insights on the interactions between them by selecting specific amino-acids that are involved in the interaction and characterizing their polarities. This new approach proposed by the MaX scientists is complementary to the docking methods used up to now and based on in-silico research of the inhibitor. Biological systems are naturally composed of fragments such as amino-acids in proteins or nitrogenous bases in DNA.
With this approach, it is possible to evaluate whether the amino acid-based fragmentation is consistent with the electronic structure resulting from the QM computation. This is an important indicator for the end-user, as it enables to evaluate the quality of the information associated with a given fragment. Then, QM observables on the system’s fragments can be obtained, which are based on a population analysis of electronic density of the system, projected on the amino-acid.
A novelty that this approach enables is the possibility of quantifying the strength of the chemical interaction between the different fragments. It is possible to select a target region and identify which fragments of the systems interact with this region by sharing electrons with it.
“We can reconstruct the fragmentation of the system in such a way as to focus on an active site in a specific portion of the protein”, says Luigi Genovese from CEA (Commissariat à l’énergie atomique et aux énergies alternatives) who is heading Max’s efforts on this topic. “We think this modelling approach could inform efforts in protein design by granting access to variables otherwise impervious to observation.”
Improving drug design and biosensors
 The project E-CAM supports HPC simulations in industry and academia through software development, training and discussion in simulation and modeling. Project members are currently following two approaches to add to the research on the corona virus.
The project E-CAM supports HPC simulations in industry and academia through software development, training and discussion in simulation and modeling. Project members are currently following two approaches to add to the research on the corona virus.
Firstly, the SARS-CoV-2 virus that causes COVID-19 uses a main protease to be functional. One of the drug targets currently under investigation is an inhibitor for this protease. While efforts on simulations of binding stability and dynamics are being conducted, not much is known of the dynamical transitions of the binding-unbinding reaction. Yet, this knowledge is crucial for improved drug design. E-CAM aims to shed light on these transitions, using a software package developed by project teams at the University of Amsterdam and the Ecole Normale Superieure in Lyon.
Secondly, E-CAM contributes to the development of the software required to design a protein-based sensor for the quick detection of COVID-19. The sensor, developed at the partner University College Dublin with the initial purpose to target influenza, is now being adapted to SARS-CoV-2. This adaptation needs DNA sequences as an input for suitable antibody-epitope pairs. High-performance computing is required to identify these DNA sequences to design and simulate the sensors prior to their expression in cell lines, purification and validation.
Studying COVID-19 infections on the cell level
 The project PerMedCoE aims to optimise codes for cell-level simulations in high-performance computing, and to bridge the gap between organ and molecular simulations. The project started in October 2020.
The project PerMedCoE aims to optimise codes for cell-level simulations in high-performance computing, and to bridge the gap between organ and molecular simulations. The project started in October 2020.
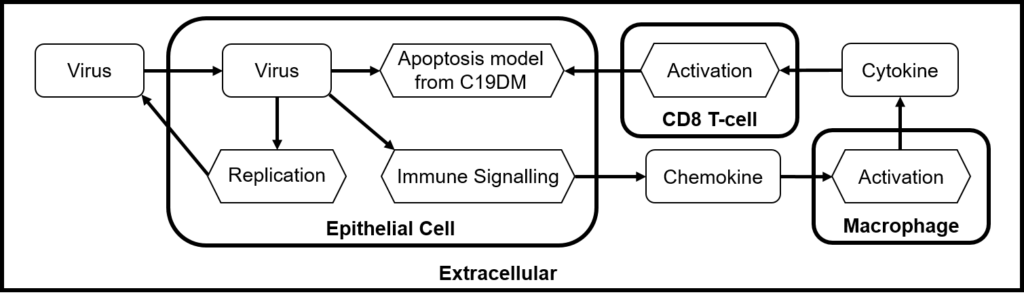
“Multiscale modelling frameworks prove useful in integrating mechanisms that have very different time and space scales, as in the study of viral infection, human host cell demise and immune cells response. Our goal is to provide such a multiscale modelling framework that includes infection mechanisms, virus propagation and detailed signalling pathways,” says Alfonso Valencia, PerMedCoE project coordinator at the Barcelona Supercomputing Center.
The project team has developed a use case that focusses on studying COVID-19 infections using single-cell data. The work was presented to the research community at a specialized virtual conference in November, the Disease Map Community Meeting. “This use case is a priority in the first months of the project”, says Valencia.
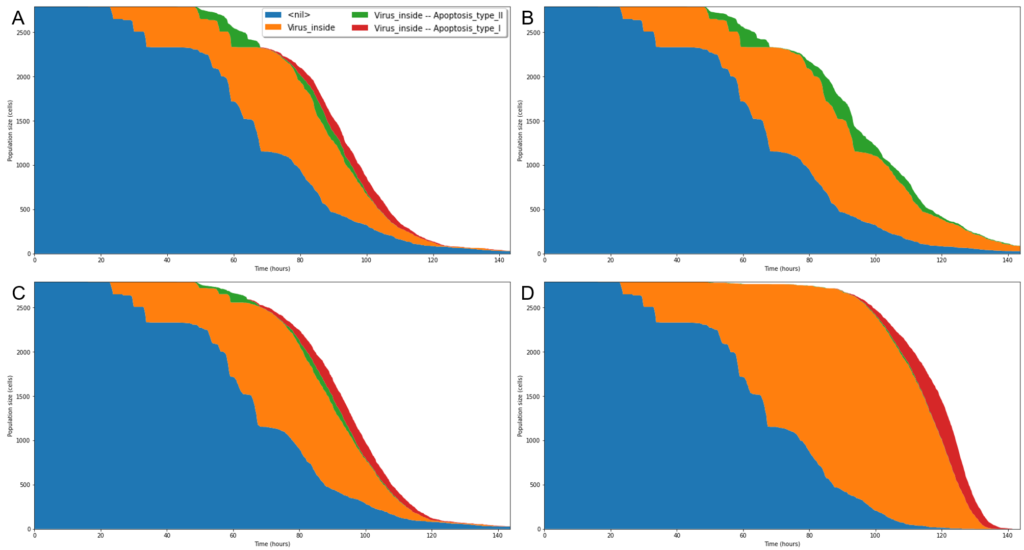
On the technical level, disease maps networks will be converted to models of COVID-19 and human cells from the lung epithelium and the immune system. Then, the team will use omics data to personalise models of different patients’ groups, differentiated for example by age or gender. These data-tailored models will then be incorporated into a COVID-focussed version of the open source cell-level simulator PhysiCell.
Supporting policy makers and governments
 The HiDALGO project focusses on modelling and simulating the complex processes which arise in connection with major global challenges. The researchers have developed the Flu and Coronavirus Simulator (FACS) with the objective to support decision makers to provide an appropriate response to the current pandemic situation taking into account health and care capabilities.
The HiDALGO project focusses on modelling and simulating the complex processes which arise in connection with major global challenges. The researchers have developed the Flu and Coronavirus Simulator (FACS) with the objective to support decision makers to provide an appropriate response to the current pandemic situation taking into account health and care capabilities.
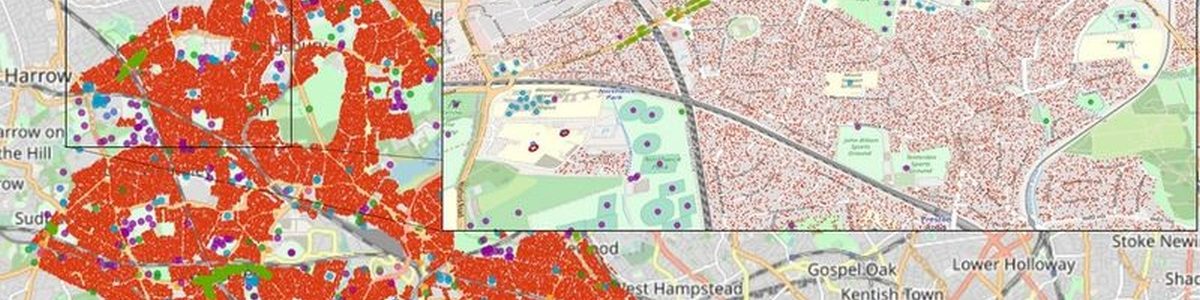
FACS is guided by the outcomes of SEIR (Susceptible-Exposed-Infectious-Recovered) models operating at national level. It uses geospatial data sources from Openstreet Map to approximate the viral spread in crowded places, while trading the potential routes to reach them.
In this way, the simulator can model the COVID-19 spread at local level to provide estimations of infections and hospital arrivals, given a range of public health interventions, going from no interventions to lockdowns. Public authorities can use the results of the simulations to identify peaks of contagion, set appropriate measures to reduce spread and provide necessary means to hospitals to prevent collapses. “FACS has enabled us to forecast the spread of COVID-19 in regions such as the London Borough of Brent. These forecasts have helped local National Health Service Trusts to more effectively plan out health and care services in response to the pandemic.” says Derek Groen from the HiDALGO project partner Brunel University London.
Wired: Why London’s Tier 2 lockdown couldn’t be borough by borough
The Guardian: As other cities go into lockdown, why isn’t London having a second wave?
HiPEAC Magazine: Simulating COVID-19 andflu spread using HiDALGO
HiDALGO website: Pandemic Use Case
HiDALGO establishes a local flu and coronavirus detector
Cordis – EU research results: London calling: Simulation tool predicts second wave of COVID-19
 EXCELLERAT is a project that is usually focussing on supercomputing applications in the area of engineering. Nevertheless, a group of researchers from EXCELLERAT’s consortium partner SSC-Services GmbH, an IT service provider in Böblingen, Germany and the High-Performance Computing Center Stuttgart (HLRS) are also providing measures to contain the pandemic by supporting the German Federal Institute for Population Research (Bundesinstitut für Bevölkerungsforschung, BiB).
EXCELLERAT is a project that is usually focussing on supercomputing applications in the area of engineering. Nevertheless, a group of researchers from EXCELLERAT’s consortium partner SSC-Services GmbH, an IT service provider in Böblingen, Germany and the High-Performance Computing Center Stuttgart (HLRS) are also providing measures to contain the pandemic by supporting the German Federal Institute for Population Research (Bundesinstitut für Bevölkerungsforschung, BiB).
The scientists have developed an intelligent data transfer platform, which enables the BiB to upload data, perform computing-heavy simulations on the HLRS’ supercomputer Hawk, and download the results. The platform supports the work of BiB researchers in predicting the demand for intensive care units during the COVID-19 pandemic. “Nowadays, organisations face various issues while dealing with HPC calculations, HPC in general or even the access to HPC resources,” said Janik Schüssler, project manager at SSC Services. “In many cases, calculations are too complex and users do not have the required know-how with HPC technologies. This is the challenge that we have taken on. The BiB’s researchers had to access HLRS’s Hawk in a very complex way. With the help of our new platform, they can easily access Hawk from anywhere and run their simulations remotely.”
“This platform is part of EXCELLERAT’s overall strategy and tools development, which not only addresses the simulation part of engineering workflows, but provides users the necessary means to optimise their work”, said Bastian Koller, Project Coordinator of EXCELLERAT and HLRS’s Managing Director. “Extending the applicability of this platform to further use cases outside of the engineering domain is a huge benefit and increases the impact of the work performed in EXCELLERAT.”
ETP4HPC handbook 2020 released
The 2020 edition of the ETP4HPC Handbook of HPC projects is available. It offers a comprehensive overview over the European HPC landscape that currently consists of around 50 active projects and initiatives. Amongst these are the 14 Centres of Excellence and FocusCoE, that are also represented in this edition of the handbook.

Press release: new CoE for Personalized Medicine launched
October 14, 2020

Coordinated by the Barcelona Supercomputing Center (BSC) and funded by the European Commission, the recently launched HPC centre of excellence PerMedCoE will optimise codes for cell-level simulations in HPC/Exascale and bridge the gap between organ and molecular simulations.
>> Initial press release [PDF]