- Homepage
- >
- Search Page
- > Use Case
- >
BioExcel: Electronic Interaction Phenomena: Proton Dynamics and Fluorescent Proteins
Short description
Proton Dynamics
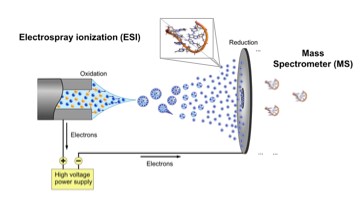
Mass spectrometry has revolutionized proteomics, i.e. the investigation of the myriad of protein/protein interactions in the cell. By using a very small sample content of proteins (usually in the micromolar concentration), a powerful implementation of the technique, so-called ionization/ion mobility mass spectrometry (ESI/IM-MS), can measure stoichiometry, shape and subunit architecture of protein and protein complexes in the gas phase.
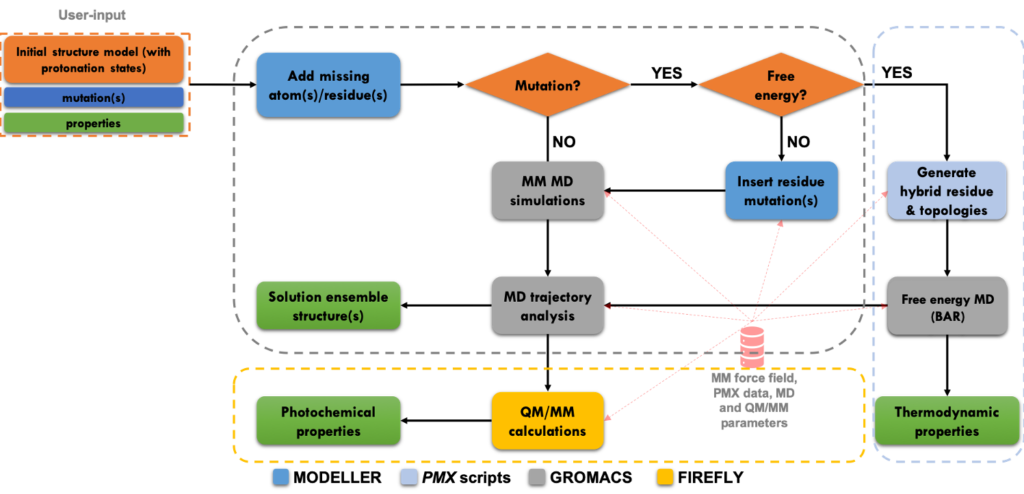
Fluorescent proteins are the backbone of many high-resolution microscopy techniques. To improve resolution, these proteins may require optimization for the specific conditions under which the imaging is carried out. While directed evolution works well if a single property needs to be optimized, optimizing multiple properties simultaneously remains challenging. Furthermore, this approach provides only limited physical insights into the process that is the target of the optimization. In contrast, computational chemistry provides a route to obtain such insights from first principles, but because such calculations typically require a high level of expertise, automatic workflows for in silico screening of fluorescent proteins are not yet generally available. To overcome this limitation we developed a user-friendly workflow for automatically computing the most relevant properties of fluorescent protein mutants based on established atomistic molecular dynamics models.
Fluorescent proteins are the backbone of many high-resolution microscopy techniques. To improve resolution, these proteins may require optimization for the specific conditions under which the imaging is carried out. While directed evolution works well if a single property needs to be optimized, optimizing multiple properties simultaneously remains challenging. Furthermore, this approach provides only limited physical insights into the process that is the target of the optimization. In contrast, computational chemistry provides a route to obtain such insights from first principles, but because such calculations typically require a high level of expertise, automatic workflows for in silico screening of fluorescent proteins are not yet generally available. To overcome this limitation we developed a user-friendly workflow for automatically computing the most relevant properties of fluorescent protein mutants based on established atomistic molecular dynamics models.
Results & Achievements
Proton Dynamics
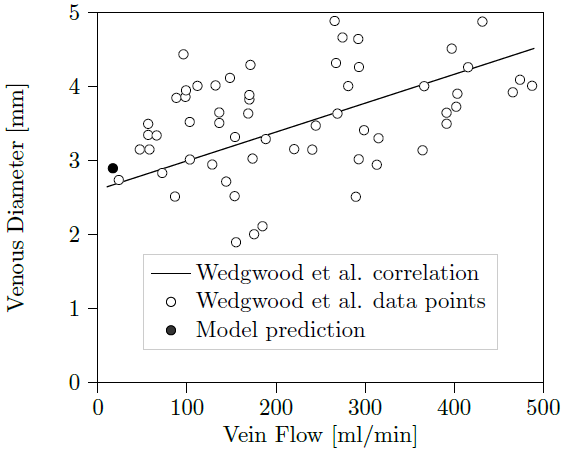
The project is in progress. It has started in December 2020. So far, we have investigated the nucleotide in water. We found a stable hairpin structure consisting of a short B-DNA segment and a d(GpApAp) triloop. Labile contact ion pairs were observed between the heptanucleotide backbone and ammonium ions, which are important for proton dynamics upon desolvation. These calculations are used as a starting point for QM/MM simulations and will be afterwards complemented by investigation of the biomolecule in the gas phase.
Fluorescent Proteins
To illustrate what can be done with the workflow, we have computed the absorption and fluorescence spectra, as well as the folding and dimerization free energies for five variants of Aequorea victoria Green Fluorescent Protein. Other important properties, such as excited-state reactivity, can be computed as well, but require significantly more computational resources. To demonstrate also this aspect of our workflow, we computed an excited state trajectory of the enhanced GFP (EGFP) variant. Because computational power, as well as the accuracy of simulation models, are expected to improve further, we anticipate that our workflow can eventually provide the fluorescence microscopy community with a powerful and predictive tool for tailoring fluorescent proteins to the specific conditions of an experiment.
Objectives
Proton Dynamics
We will use massively parallel hybrid QM/MM approaches to investigate proton dynamics during the desolvation process of biomolecules during mass-spec experiments. The approaches include the previously developed interface (coupling GROMACS to CPMD) to the new interface (coupling GROMACS to CP2K) developed by BioExcel. We will focus on the heptanucleotide d(GpCpGpApApGpC). Comparison with the available experimental data will allow us to establish the accuracy of the methods used. This project will illustrate computational scaling, performance, and efficiency of BioExcel’s QM/MM codes, run on a highly relevant biological problem.
Fluorescent Proteins
We developed a user-friendly workflow for automatically computing the most relevant properties of fluorescent protein mutants based on established atomistic molecular dynamics models. In particular, the workflow combines classical molecular dynamics, with free energy computations and hybrid quantum mechanics / molecular mechanics (QM/MM) simulations to compute the effects of mutations on the key properties of these proteins, including absorption and emission wavelengths, folding free energy, oligomerization affinities, fluorescence and switching quantum yields.
Performance Portability on HPC Accelerator Architectures with Modern Techniques:
The ParFlow Blueprint
A Use Case by 
Short description
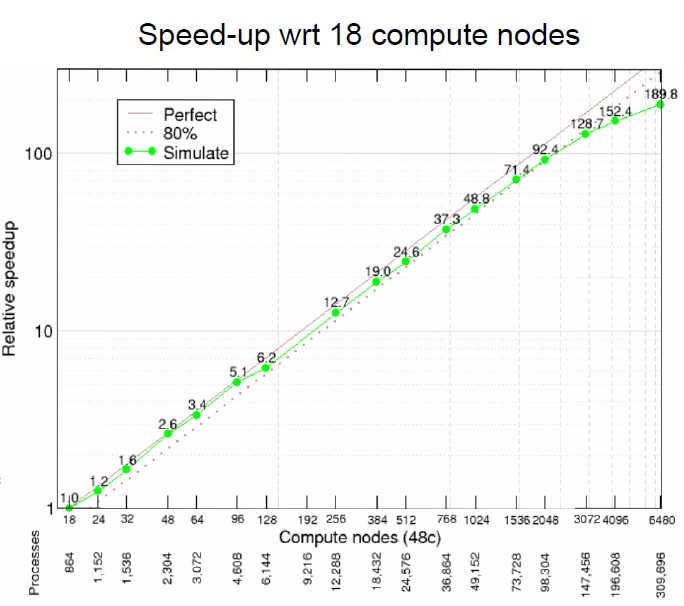
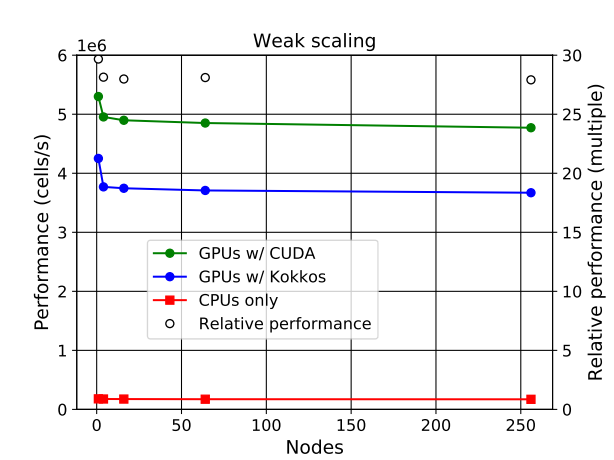
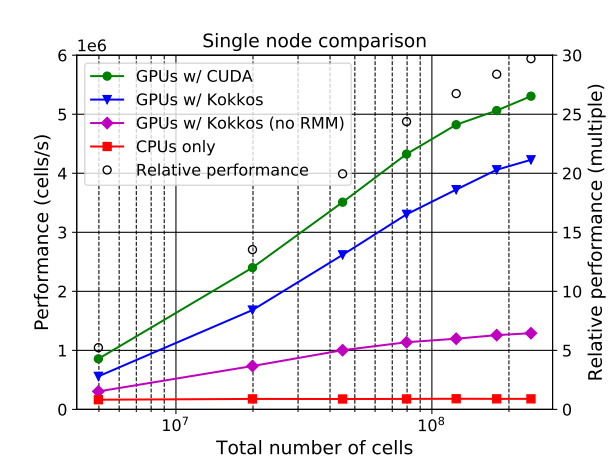
Rapidly changing heterogeneous supercomputer architectures pose a great challenge to many scientific communities trying to leverage the latest technology in high-performance computing. Software development techniques that simultaneously result in good performance and developer productivity while keeping the codebase adaptable and well maintainable in the long-term are of high importance. ParFlow, a widely used hydrologic model based on C, achieves these attributes by using Unified Memory with a pool allocator and abstracting the architecture-dependent code in preprocessor macros (ParFlow eDSL). The implementation results in very good weak scaling with up to 26x speedup from the NVIDIA A100 GPUs over hundreds of nodes each containing 4 GPUs on the new JUWELS Booster system at the Jülich Supercomputing Centre.
Results & Achievements
The GPU support have been successfully implemented for ParFlow through using either CUDA or Kokkos (these are two separate lightweight implementations that can be used separately from each other). Especially the implementation of Kokkos constitutes a big leap in performance portability. Up to 26x speedup is achieved when using GPUs, versus using only CPUs.
Objectives
Objectives are to achieve performance portability developing and applying the ParFlow eDSL including support for coupled simulations where ParFlow is used in combination with other independently developed terrestrial models such as land-surface and atmospheric models. Multi-vendor support (i.e. performance portability) is preferred to guarantee compatibility with different supercomputer architectures.
Technologies
ParFlow, CUDA, Kokkos
Use Case Owner
FZ-Juelich IBG3
Collaborating Institutions
LLNL, FZ-Juelich
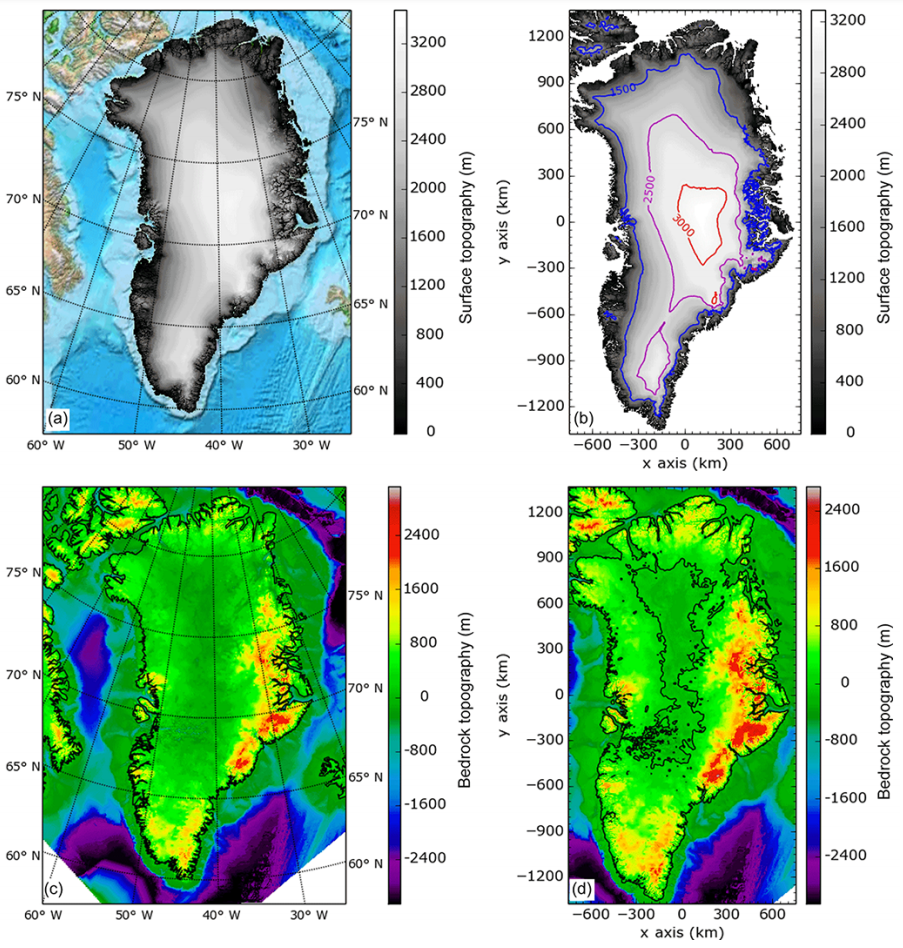
OBLIMAP ice sheet model coupler parallelization and optimization
Short description
Within the ESiWACE2 project, we have parallelized and optimized OBLIMAP. OBLIMAP is a climate model - ice sheet model coupler that can be used for offline and online coupling with embeddable mapping routines. In order to anticipate future demand concerning higher resolution and/or adaptive mesh applications, a parallel implementation of OBLIMAP's fortran code with MPI has been developed. The data-intense nature of this mapping task required a shared memory approach across the processors per compute node in order to prevent the node memory from being the limiting bottleneck. Moreover, the current parallel implementation allows multi-node scaling and includes parallel NetCDF IO in addition to loop optimizations.
Results & Achievements
Results show that the new parallel implementation offers better performance and scales well. On a single node, the shared memory approach allows now to use all the available cores, up to 128 cores in our experiments of the Antarctica 20x20km test case where the original code was limited to 64 cores on this high-end node, and was even limited to 8 cores on moderate platforms. The multi-node parallelization yields for the Greenland 2x2km test case a speedup of 4.4 on 4 high-end compute nodes equipped with 128 cores each when compared to the original code, which was able to run only on 1 node. This paves the way to establishing OBLIMAP as a candidate ice sheet coupling library for large-scale, high-resolution climate modeling.
Objectives
The goal of the project is firstly to reduce the memory footprint of the code by improving its distribution over parallel tasks, and secondly to resolve the I/O bottleneck by implementing parallel reading and writing. This will improve the intra-node scaling of OBLIMAP-2.0 by using all the cores of a node. A second step will be the extension of the parallelization scheme to support inter-node execution. This work will establish OBLIMAP-2.0 as a candidate ice coupling library for large-scale, high-resolution climate models.
Collaborating Institutions
KNMI
Atos
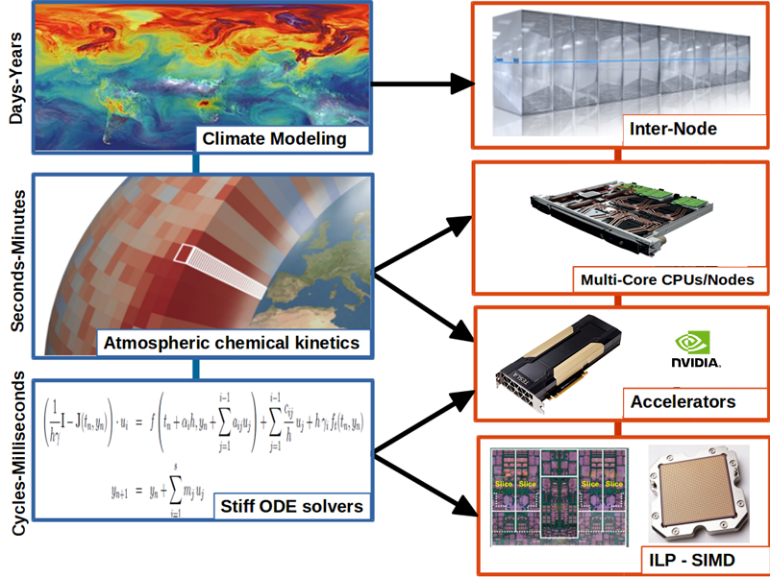
GPU Optimizations for Atmospheric Chemical Kinetics
Short description
Within the ESiWACE2 project, open HPC services to the Earth system modelling community in Europe provide guidance, engineering, and advice to support exascale preparations for weather and climate models. ESiWACE2 aims to improve model efficiency and to enable porting models to existing and upcoming HPC systems in Europe, with a focus on accelerators such as GPUs. In this context, through a collaboration between Cyprus Institute, Atos, NLeSC and also with the participation of Forschungszentrum Jülich, the ECHAM/MESSy Atmospheric Chemistry model EMAC has been optimized. EMAC describes chemical interactions in the atmosphere, including sources from ocean biochemistry, land processes and anthropogenic emissions. This computationally intensive code was ported in the past to GPUs using CUDA to achieve speedups of a factor of 5-10. The application had a high memory footprint, which precluded handling very large problems such as more complex chemistry.
Results & Achievements
Thanks to a series of optimizations to alleviate stack memory overflow issues, the performance of GPU computational kernels in atmospheric chemical kinetics model simulations has been improved. Overall, the memory consumption of EMAC has been reduced by a factor of 5, allowing a time to solution speedup of 1.82 on a benchmark representative of a real-world application, simulating one model month.As a result, we obtained a 23% time reduction with respect to the GPU-only execution. In practice, this represents a performance boost equivalent to attaching an additional GPU per node and thus a much more efficient exploitation of the resources.
Objectives
The goal of the project service was to reduce the memory footprint of the EMAC code in the GPU device, thereby allowing more MPI tasks to be run concurrently on the same hardware. This allowed the model to be optimized reaching high performance for current and future GPU technologies and later to extend its computational capability, enabling it to handle chemistry that is an order of magnitude more complex, such as the Mainz Organic Mechanism (MOM).
Technologies
EMAC (ECHAM/MESSy) code
CUDA
MPI
NVIDIA GPU accelerator
Atos BullSequana XH2000 supercomputer
In silico trials for effects of COVID-19 drugs on heart
Short description
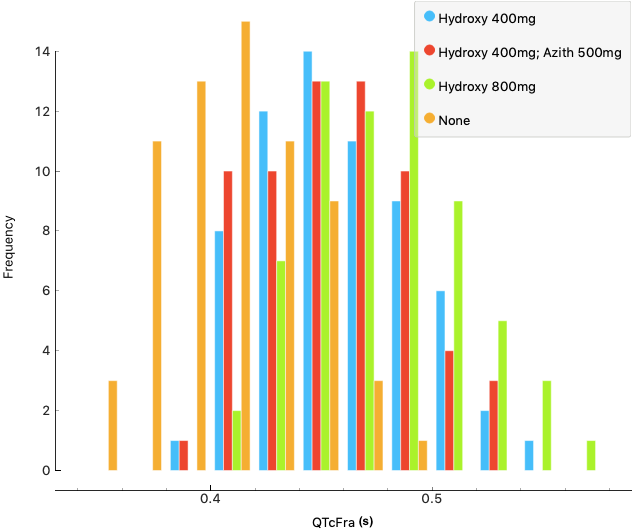
Treatment of COVID-19 identified two potentially effective drugs, azithromycin and hydroxychloroquine, however, these were known to have proarrhythmic effects (disruption of the rhythm of the heart) especially in those with underlying heart conditions. Little was known about the effect of these drugs on the arrhythmic risk to patients who receive it as a COVID-19 therapy or the effect on the heart of using the drugs in combination.
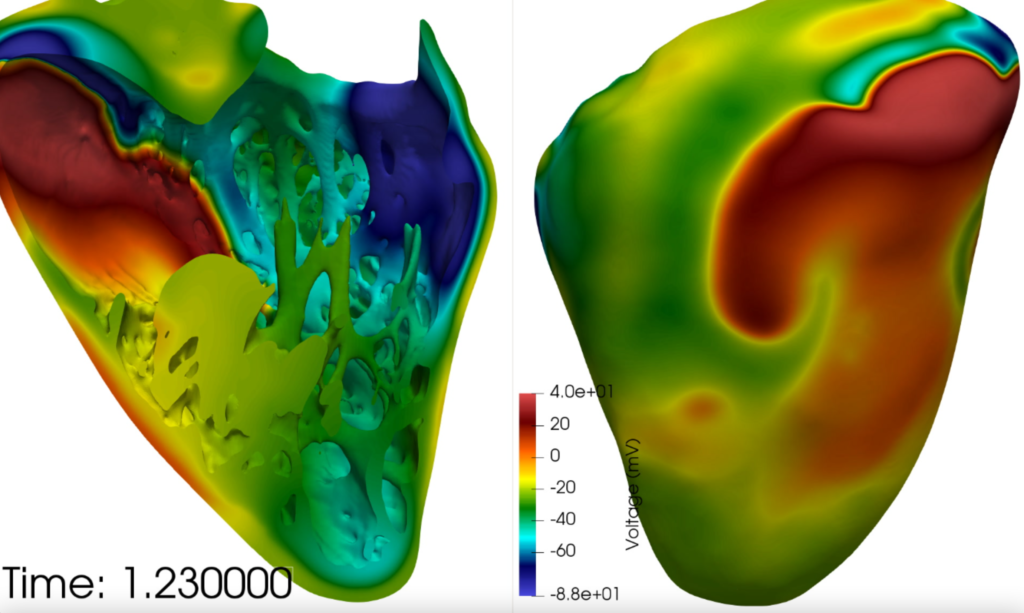
Barcelona Supercomputing Centre (BSC) conducted research in collaboration with ELEM Biotech (ELEM) to assess the impact of the drugs on the human heart, to provide guidance for clinicians on dosages and risks. Alya Red is used in their research, a multiscale model developed by BSC which performs cardiac electro-mechanics simulations, from tissue to organ level. This type of computational modelling can provide a unique tool to develop models for the function of the heart and assess the cardiotoxicity of drugs to a high level of detail, without the need for lengthy and expensive clinical research.
The state-of-the-art models being employed and developed will also enable them to study the varied effects between groups of individuals and look at the comorbidities for such things as gender, ischemia, metabolite imbalances, and structural diseases.
Results & Achievements
Early results from the BSC/ELEM models have already identified how a conduction block may occur as an effect from action potential duration lengthening from the use of chloroquine. The arrhythmic risk was assessed during a stress test on the population (at elevated heart around 150 bpm) and with a variety of doses of the medication.
The results obtained on the human population (QTc interval duration and arrhythmic risk) were compared to clinical trials performed on COVID-19 (SARS-CoV-2) patients. An in-silico cardiac population was capable of providing remarkably similar percentages of subjects at risk, as were identified in the clinic. Furthermore, the virtual population allowed the identification of phenotypes at risk of drug-induced arrhythmias.
Objectives
- assess the impact of the drugs on a human heart population
- provide guidance for clinicians on dosages and risks
- study the varied effects between groups of individuals.
- analyse the effect of the antimalarial drugs on a range of simulated human hearts with a variety of comorbidities which may be present in an infected population.
Collaborating Institutions
Visible Heart Laboratories
University of Minnesota
CT2S - Computer Tomography to Strength
Short description
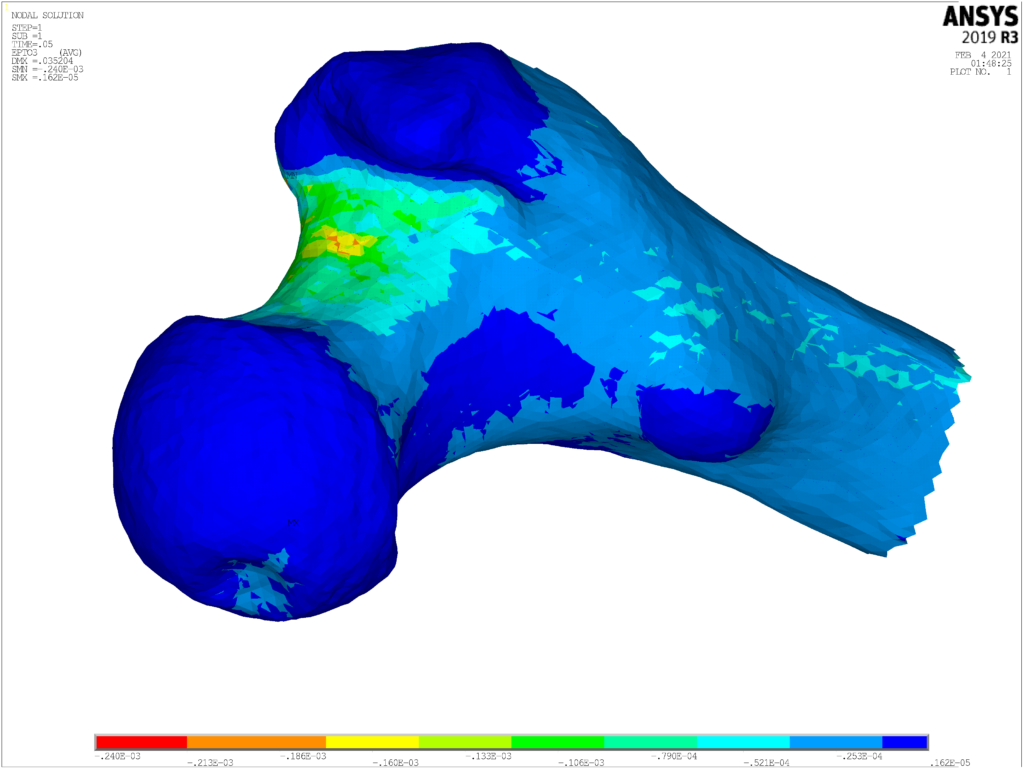
The CT2S (Computer Tomography to Strength) application was designed to predict the risk of hip fracture at the time of a CT scan, creating a patient-specific finite-element model of the femur. The CT2S pipeline comprises femur geometry segmentation, high-quality 3D mesh generation, element-wise material property mapping, and eventually finite-element model simulation. By using a disease progression model, this is now being extended to assess the Absolute Risk of Fracture up to 10 years (ARF10). In parallel with this, a solution known as BoneStrength is being developed (based on algorithms in CT2S and ARF10) to build an in silico trial application for bone-related drugs and treatments. Currently, the BoneStrength pipeline is being used on a cohort of 1000 digital patients to simulate the effect of an osteoporosis drug.
Results & Achievements
- M. Taylor, M. Viceconti, P. Bhattacharya, X. Li 2021. “Finite element analysis informed var-iable selection for femoral fracture risk predic-tion”, Journal of the Mechanical Behavior of Biomedical Materials, in press.
- C. Winsor, X. Li, M. Qasim, C.R. Henak, P.J. Pickhardt, H. Ploeg, M. Viceconti 2021. “Eval-uation of patient tissue selection methods for deriving equivalent density calibration for femo-ral bone quantitative CT analyses”, Bone, 143, 115759.
- Z. Altai, E. Montefiori, B. van Veen, M. A. Paggiosi, E. V. McCloskey, M. Viceconti, C. Mazzà, X. Li 2021. “Femoral neck strain pre-diction during level walking using a combined musculoskeletal and finite element model ap-proach”, PLoS ONE, 68. https://doi.org/10.1371/journal.pone.0245121
Objectives
- To develop an efficient digital twin solution for the estimation of long bone fracture risk in elderly patients (CT2S solution)
- To not only be able to evaluate the bone strength at present, but also predict the changes in the next 10 years (ARF10 solution)
- To extend the digital twin solution to predict other types of fracture, such as the vertebral
- To enable the running of large scale in silico clinical trials (BoneStrength solution, with a current target for bone treatments)
- To port these solutions on multiple HPC platforms across Europe and solve potential scalability issues
Collaborating Institutions
University of Bologna
University of Sheffield
Strong scaling performance for human scale blood-flow modelling
Short description
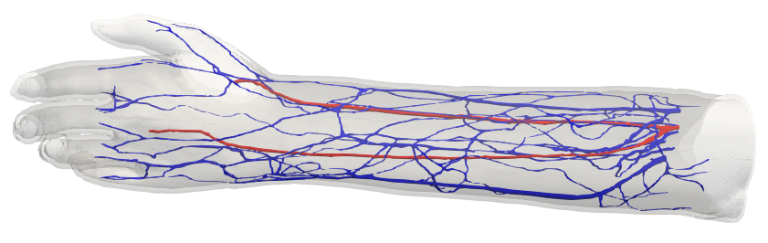
To get the best out of future exascale machines, current codes must be able to demonstrate good scaling performance on current machines. The HemeLB code has demonstrated such characteristics to full machine scale on SuperMUC-NG. However, this performance must be aligned with the ability to solve practical problems. We have developed and deployed a self-coupled version of HemeLB that allows us to simultaneously study flows in linked arterial and venous vasculatures in 3D. In this use case, we look towards the application of flow in an arteriovenous fistula. A fistula is created in patients with kidney failure to increase flow in a chosen vein in order to provide an access point for dialysis treatment.
Results & Achievements
In collaboration with POP CoE, we were able to demonstrate HemeLB’s capacity for strong scaling behaviour up to the full production partition of SuperMUC-NG (>300,000 cores) whilst using a non-trivial vascular domain. This highlighted several challenges of running simulations at scale and also identified avenues for us to improve the performance of the HemeLB code.
We’ve also run self-coupled simulations on personalised 3D arteries and veins of the left forearm with and without an arteriovenous fistula being created. The initial flow from our modified model showed good agreement with that seen in a clinical study.
Objectives
The first objective of this use case was to demonstrate and assess the strong scaling performance of HemeLB to the largest core counts possible. This was to enable us to evaluate current performance and identify improvements for future exascale machines. The second main objective was to demonstrate the ability of HemeLB to utilise this performance to study flows on human-scale vasculatures. The combination of both aspects will be essential to enabling the creation of a virtual human able to simulate the specific physiology of an individual for diagnostic purposes and evaluation of potential treatments.
Fuel atomization and evaporation in practical applications
Short description
Spray modelling for liquid fuels is another milestone to be covered for a successful engine simulation. Since the quality of atomization, evaporation and dispersion predictions directly affect the overall phenomena developing downstream, its proper modelling has a first-order impact on the whole results. In the context of LES and DNS, primary atomization can be described by deforming the mesh at the interface or by transporting Eulerian fields from which the interface is reconstructed a posteriori. After this process, the secondary atomization occurs in which the large droplets and ligaments are broken into small droplets. Although secondary atomization can be formulated in the frame of Eulerian transport equations, the size of the cells determines the minimum length that can be solved making attractive the use of the alternative Lagrangian approach. The behaviour of the droplets and their evolution have been object of an intense study over the years and, although a fruitful research has been carried out in this topic, much effort is still needed in order to develop comprehensive models for the primary atomization that can lead to physically accurate formulations for application to spray flames.
Objectives
It is considered a priority to contribute to the knowledge of liquid fuel injection and atomization due to its crucial influence on the combustion process and pollutants formation. This demonstrator includes the study of primary and secondary breakup, and the influence of heat conduction and droplet heating on the evaporation rates prior combustion takes place. The final objective will be the study of reacting sprays at relevant engine conditions.

Plasma assisted combustion
Short description
Plasma-assisted combustion has recently gained renewed interest in the context of lean burning. Although lean combustion reduces the burn gas temperature and CO/CO2 content it leads, however, to less stable flames, and more difficult ignition. Short plasma discharges may counteract these effects, at a very low additional energy cost. In particular, Nanosecond Repetitively Pulse (NRP) plasma discharges have been proven to be efficient actuators to alter flame dynamics and facilitate flame stabilization while modifying the burning velocity. First studies on the impact of NRP discharges on combustion focused on kinetic mechanisms and gas heating processes. Different models have been developed in 0D, but no kinetic scheme for plasma-assisted combustion has been proposed so far. More recently 1D and 2D simulations have been conducted. Very few studies included a self-consistent simulation of the NRP discharge for combustion applications, although it was shown that thermal and chemical effects of the NRP discharge on ignition are of the same order. Because of their high computational cost, detailed simulations of 3D real cases have never been performed and only simplified formulations for the plasma effects have been used.
Objectives
The application of plasma in combustion simulations provides an unprecedented opportunity for combustion and emission control thanks to its capability to produce heat, active radical species and modify the transport properties of the mixture. This demonstrator is focused on the study of plasma-assisted combustion by Nanosecond Repetitively Pulsed (NRP) discharges in order to control the formation of combustion instabilities and pollutant formation.

Detailed chemistry DNS calculation of turbulent hydrogen and hydrogen-blends combustion
Short description
A block of actions to decarbonize the EU is the use of low carbon content fuels, comprising natural gas and hydrogen blends. Regarding natural gas, its low carbon content leads to relatively low CO2 emissions while its higher resistance to knock compared to gasoline allows to achieve higher compression ratios and hence higher efficiencies. In spite of these benefits, such resilience may provoke a non-stable operation of the engine that can be avoided by the addition of hydrogen which expands the flammability region. The effects of hydrogen addition consist of the increase of the laminar flame speed and the induction of preferential diffusion which can result in thermo-diffusive instabilities. Although some works have analysed these phenomena, more effort has to be devoted. Finally, hydrogen blends and syngas, fuels with high hydrogen content (HHC), are other alternatives to reduce green house gas emissions that will also be investigated. Analogously, to natural gas blend with hydrogen, HHC fuels show thermo-diffusive instabilities which may lead to an unstable combustion process. Although some works have addressed these issues, there is still a lack of knowledge to be covered. In this context, LES and DNS come up as powerful techniques set in the context of HPC that can shed light in many open questions.
Objectives
The use case focusses on the study of thermo-diffusive instabilities in turbulent lean hydrogen flames and its effects on burning velocities, unstable combustion and noise. The effect of preferential diffusion will also be investigated due to its influence on equivalence ratio fluctuations and eventually on the local burning velocity. This work will be extended to syngas and high hydrogen content (HHC) fuels.