POP and PerMed Centers of Excellence are Getting Cell-Level Simulations Ready for Exascale
Success story # Highlights:
- Industry sector: Computational biology
- Key codes used: PhysiCell
- Keywords:
- Memory management
- Cell-cell interaction simulation
- OpenMP
- Good practice
Organisations & Codes Involved:



Technical Challenge:
One of PerMedCoE’s use cases is the study of tumour evolution based on single-cell omics and imaging using PhysiCell. In order to simulate such large-scale problems that replicate real-world tumours, High Performance Computing (HPC) is essential. However, memory usage presents a challenge in HPC architectures and is one of the obstacles to optimizing simulations for running at Exascale.
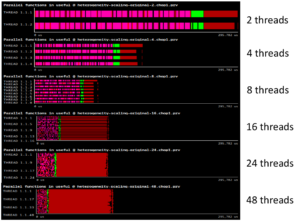
With a high number of threads computing in parallel, performance can be degraded because of concurrent memory allocations and deallocations. We observe that as the number of threads goes up, runtime is not reduced as expected. This is not a memory-bound problem, but a memory management one.
Solution:
Following the POP methodology and using the BSC tools (Extrae, Paraver, and BasicAnalysis), we determine that an overloading of a specific implementation of C++ operators is causing a high number of concurrent memory allocations and deallocations, causing the memory management library to perform synchronization and “steal” cycles of CPU from the running application.
The proposal from POP is to avoid the overloading of operators that need allocation and deallocation of data structures. However, achieving this implies a major code change. To demonstrate the potential of the suggestion without doing the major code change, we suggest using an external library to improve the memory management: Jemalloc.
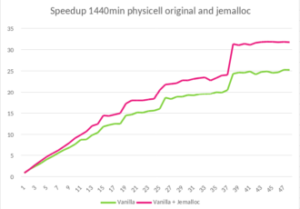
Jemalloc is “a general purpose malloc implementation that emphasizes fragmentation avoidance and scalable concurrency support” [1]. It can be integrated easily into any code by preloading the library, with LD_PRELOAD in Linux for example. After applying this solution to PhysiCell, we execute the same experiment and obtain a 1.45x speed up when using 48 OpenMP threads.
[1] cited from Jemalloc website. http://jemalloc.net
Business impact:
As the only transversal CoE, one of POP’s objectives is to advise and help the other HPC CoEs prepare their codes for the Exascale. This collaboration between POP and PerMed is a good example of the potential of these kinds of partnerships between CoEs.
POP provides the performance analysis expertise and tools, the experience of hundreds of codes analysed, and the best practices gathered from those analyses. PerMedCoE bring state of the art use cases and real-world problems to be solved. Together, they improve the performance and efficiency of cell-level agent-based simulation software: in this case PhysiCell.
One of the goals of PerMedCoE is the scaling-up of four different tools that address different types of simulations in personalized medicine and that were coded in different languages (C++, R, python, julia). These tools are being re-factored to scale up to Exascale. In this scaling-up, audits such as the ones POP can offer are essential to evaluate past developments and guide future ones.
Currently, only one of PerMed’s tools is able to use several nodes to run a single simulation, whereas the rest of the tools can only use all of the processors of a single node. Our scaling-up strategy is a heterogeneous one. We are implementing MPI on some tools, re-factoring others to other languages that can ease the use of HPC clusters such as Julia, or even targeting “many-task computing“ paradigms like in the case of model fitting.
The main motivation to have HPC versions of these simulation tools is to be able to simulate bigger, more complex cell-level agent-based models. Current models can obtain up to 10^6 cells, but it has been proven that most of the problems addressed in computational biology (cancerogenesis, cell lines growth, COVID-19 infection) usually target from 10^9 to 10^12 cells. In addition, most of these current simulations consider an over-simplified environment that is nowhere close to a real-life scenario. We aim to have complex multi-scale simulation frameworks that target these bigger, more complex simulations.
Benefits:
- Speedup of 1.45x on runtime with 48 threads in PhysiCell execution
- A good practice for High Performance C++ applications exported
- This behaviour in memory management systems will be detected more easily in future analysis



